Probiotic memiRNAs: New Insights for Infection Control
The Role of Prokaryotic (Probiotic) Produced Megamicro RNAs (memiRNAs) to Prevent or Cure Viral and Nosocomial Infections, Certain Cancers, in Conjunction with Probiotic Induced Eukaryotic Micro RNAs (miRNAs)
Dr. Malireddy S. Reddy, BVSc (DVM)., MS., Ph.D1
OPEN ACCESS
PUBLISHED: 31 December 2024
CITATION: Reddy, MS., 2024. The Role of Prokaryotic (Probiotic) Produced Megamicro RNAs (memiRNAs) to Prevent or Cure Viral and Nosocomial Infections, Certain Cancers, in Conjunction with Probiotic Induced Eukaryotic Micro RNAs (miRNAs). Medical Research Archives, [online] 12(12). https://doi.org/10.18103/mra.v12i12.6179
COPYRIGHT: © 2025 European Society of Medicine. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
DOI: https://doi.org/10.18103/mra.v12i12.6179
ISSN 2375-1924
ABSTRACT
In 2021 US Patent # 11,077,052 B1, entitled “Selected Multiphase Treatment for Coronavirus Respiratory Infections” had been issued. The main theme of the successful, practical and novel patent was the use of multiple mixed strain probiotics along with their immunomodulins to prevent or treat Covid-19 infections during the Global Pandemic. The patented invention was accomplished through use of a selective probiotic based Mouth Wash, Nasal Spurge, Nasal Inhalation, and Oral Administration of a Liposomal Preparation. Extensive clinical trials, as outlined in the patent and published data in the peer reviewed journal of Medical Research Archives of the European Society of Medicine titled “ Mechanism of Thrombosis during Covid-19 infection due to SARS-CoV-2 virus and its variants, and a clinically proven strategy to combat with probiotics and their immunomodulins” further proved and validated the invention with regard to its efficacy as a preventative or curative aid to curb Covid-19 infections (after conducting more clinical trials). Logical explanations and partial scientific proof were presented regarding the biochemical mechanism of the multiple mixed strain probiotics and their immunomodulins on curbing Covid-19 infections. The current investigation was undertaken to study if there could be any other additional factors such as microRNAs, produced by the probiotics as part of the immunomodulins, that might be responsible for the success of this novel patented preventive or treatment modality. Multiple chronological research experiments conducted during this investigation proved that the probiotic-produced intracellular microRNAs, (mega-microRNAs), produced after they attained stationary growth phase, exhibited inhibition of some of the pathogenic bacteria. For the first time in the history of Bacteriology, it was discovered and proven that the autoinhibition of growth of the probiotic bacteria (during stationary phase) was due to the slow down or temporary stoppage of the translation of mRNA coding for the growth proteins by RNA related compounds, preferably miRNAs. The results also proved that probiotics produced microRNAs (mega-microRNAs or memiRNAs) under the influence of bacteriophage exhibited different inhibitory patterns than the ones produced without bacteriophage influence. Finally, this investigation revealed that the probiotic produced microRNAs have both inhibitory as well as stimulatory effects on bacterial growth protein translation, depending on the conditions and the circumstances under which they were produced. Since they are larger than eukaryotic microRNAs, we have named them, for the first time, as mega-microRNAs or memiRNAs. Thus, miRNA is of eukaryotic origin, whereas memiRNA is of prokaryotic origin. (For the benefit of the reader, these terms have been interchangeably used). The results also proved the partial reason for the success of the invention outlined in US Pat # 11,077,052 B1 to curb RNA viruses could be due to the combined effect of probiotic-produced mega-micro RNAs (memiRNAs) as part of the immunomodulins in conjunction with microRNAs of the host eukaryotic cells, and also the probiotic bacteria themselves.
Keywords
Eukaryotic microRNA (miRNA); Prokaryotic mega-microRNA (memiRNA); Dr. M.S. Reddy’s Multiple Mixed Strain Probiotic Therapy; Immunomodulins; COVID-19; Adjuvant Cancer Therapy; Nosocomial Infections; Streptococcus thermophilus; Lactobacillus helveticus; Bacteriophage; Protease; Amino peptidase; DNase; RNase; SARS-CoV-2; Fecal Microbiota Therapy; C. diff; MRSA; Standard Cancer Therapy; US PAT # 11,077, 052 B1; US PAT # 11,643, 641 B2; US PAT # 6,998, 700.
Introduction
Several patents and journal publications appeared during the Covid-19 Pandemic regarding the prevention and/or treatment of Covid-19. The breakthrough novel patent, US Patent # 11,077,052 B1 was an invention utilizing multiple mixed strain probiotics along with their immunomodulins to prevent or treat Covid-19 infections. This was the first biotech patent that was approved by the United States Patent Office on this subject. According to the patent the main therapeutic principle behind the invention was the immunomodulins produced by the multiple mixed strain probiotics along with live probiotics; the immunomodulins included short chain fatty acids, multiple strain and species specific bacteriocins, therapeutic peptides, hydrogen peroxide, organic acids including but not limited to Lactic, Acetic, Butyric and Propionic acids etc. However, no mention was made of the microRNAs (memiRNAs) produced by the probiotic bacteria; these end up as one of the key ingredients of the immunomodulins (or the growth end products) of the multiple mixed strain probiotics which induce the inhibition of the SARS-CoV-2 Corona virus and other secondary bacterial infections associated with Covid-19. This is done through maintaining optimal composition of the microbiota, which is essential not only to eliminate dysbiosis but also to orchestrate the immunomodulation to control a cytokine storm.
The Discovery of microRNA
In 2024, the Nobel prize in Physiology or Medicine was awarded to Victor Ambros and Gary Ruvkun for their discovery of microRNA (aka miRNA) and its function in the post-transcriptional regulation of genes in the eukaryotic cells. Victor Ambros discovered that Lin-4 gene does not encode a protein but rather produces a short non-coding RNA only 22 Nucleotides long. At the same time, Gary Ruvkun came up with a discovery that Lin-14 gene’s expression was regulated post-transcriptionally. Before their discovery, it was widely believed that the regulation of protein coding gene expression occurred primarily at the transcriptional level within the gene itself. They demonstrated that a short RNA (miRNA) in the eukaryotic cells could regulate protein synthesis post-transcriptionally by interacting with mRNA, and the first discovery of miRNA was credited to Drs. Ambros and Ruvkun. Despite this monumental discovery, microRNAs of the prokaryotic cells – specifically probiotics – has not been paid much attention.
The Definition of microRNA (aka miRNA)
MicroRNA (miRNA) is a short single-stranded RNA molecule that does not code for proteins, but instead functions to regulate the gene expression by binding to the messenger RNA (mRNA) thereby preventing mRNA from being translated into proteins and effectively silencing or degrading the target gene, acting as a “Molecule Switch” to control various cellular processes like cell growth, differentiation and apoptosis etc. in eukaryotes. These microRNAs (miRNAs) play a key role in several normal physiological functions in human biological systems, and their deregulation or abnormalities has been implicated in the pathogenesis of several diseases including cancers, autoimmune diseases, central nervous system disasters, Alzheimer disease, Rheumatoid Arthritis, and several viral and bacteriological infections etc. Although there are several research articles that have appeared on this subject with reference to eukaryotic miRNA, there is not much research done in prokaryotic bacteria, specifically on probiotics. MicroRNAs are non-coding RNAs, unlike Messenger RNA (mRNA) which codes for specific proteins. The typical mRNA molecule contains significantly more nucleotides – to the tune of hundreds to thousands – whereas eukaryotic microRNAs contain only around 21-24 nucleotides. In other words mRNA is a thousand times larger than microRNA, and its size is dictated by the gene it codes for. Eukaryotic human cells will have close to 25,000 genes in each cell. Out of the 25,000 genes it is roughly estimated that 2200 genes may code for microRNAs. Yet these micro RNAs control 60 percent of the gene expressions. Although mRNA directly codes for the proteins, other types of RNA like ribosomal RNAs (rRNA) and transfer RNA (tRNA) are considered non-coding RNAs that play structural roles in protein synthesis but do not directly encode amino acid sequences. Thus, mRNA is considered as a coding RNA. As is commonly known, mRNA serves as a gene-coding template during protein synthesis, while tRNA transports the amino acids to the ribosomes that needed to be added to the polypeptide chain. On the other hand, rRNA, in combination with proteins, forms ribosomes. In addition, there are some small interfering RNAs (siRNA) which are highly specific with one mRNA target, specifically to provide viral defense and genome stability. Considering all the above RNAs, microRNAs specifically function as endogenous gene expression regulators. MicroRNA can inhibit the translation of multiple mRNA targets from forming specific proteins. A single microRNA can inhibit several mRNAs, unlike siRNA, which can target only one mRNA. This is because of the nature of microRNAs imperfection in pairing. Generally, in eukaryotic cells, genes are copied into mRNA through transcription in the nucleus. Then the mRNA is moved out of nucleus into cytoplasm where it is translated into specific protein with the aid of tRNA and rRNA, whereas microRNAs controls gene expressions mainly by binding with the mRNA in the cell cytoplasm. Instead of being translated into protein, the marked mRNA (due to binding of microRNA) will either be destroyed and its components recycled (autophagy), or it will be preserved and translated later. In order to appreciate the novelty of the current breakthrough discovery by the readers, the following scientific information is presented, in a simplified version, with regard to the production and role of microRNAs and their functions in eukaryotic and prokaryotic cells, as we know as of now, by taking into account the established facts and hypothesis.
Biochemical mechanism of the production of Eukaryotic cell to produce microRNAs (miRNAs):
Although according to limited research, microRNAs are only produced by eukaryotic cells, yet their existence and role in prokaryotic cells cannot be ignored either. In eukaryotic cells, there are certain dedicated genes in the human genome for microRNAs. These genes do not encode for a protein, but they encode only for miRNA. However, these non-coding RNAs (miRNAs) play an important role in gene expression and the overall gene regulation. The gene expression and the final step-wise formation of the mature microRNAs and their role on disintegrating or blocking translation of mRNA in the eucaryotic cells is as follows: The first transcript which comes out of the microRNA coding gene is the Primary RNA Transcript (pri-miRNA). This pri-miRNA is further processed with the aid of endoribonucleases DROSHA and PASHA (dgcr-8) to result in pre-miRNA in the nucleus. This pre-miRNA is then transferred out of the nucleus into the cell cytoplasm with the aid of exporting-5 and RAN GTP complex. The pre-miRNA is further trimmed with the aid of DICER in the cytoplasm, thus resulting in two separate strands of mature miRNAs (miRNA). The miRNA in association with Argonaut protein developed into RNA Induced Silencing Complex (RISC). This miRNA-RISC complex binds to the messenger RNA (mRNA) to down regulate mRNA by degradation. Since mRNA has 3’UTR (3 prime Untranslated Region) and 5’ UTR at both ends, with the center part being the coding region, the miRNA-RISC complex binds mostly at 3’ UTR of mRNA and degrades it to pieces thus making it nonfunctional, from translation into a specific protein. In some instances, this miRNA-RISC complex just sits in the 3’ UTR and does not cleave the mRNA, but rather acts as a roadblock through translation interference to prevent the protein synthesis. Now let us look into the scenario in prokaryotic cells.
MicroRNA production and role in prokaryotic cells (memiRNA):
Since prokaryotic cells do not have defined nuclear membranes and enzymes DROSHA, PASHA, and even perhaps DICER, the length (dictated by number of nucleotides) of the miRNAs of the prokaryotes (memiRNAs) could be larger than the eukaryotes yet functions just like miRNAs of the eukaryotes. Prokaryotic memiRNA may downregulate the host mRNA, yet it may not degrade it totally. However, hypothetically, if they encounter foreign mRNA (viral etc.), they may downregulate the viral or foreign mRNA through degradation. Thus the trigger for the production of prokaryotic miRNA (memiRNA) can be to protect the bacterial cell from burn-out (or excess growth) or to eliminate the production of weaker mutants or to protect it from viral infections, or to induce lysogeny, or to assist in the formation of CRISPR etc. These mechanisms are not well studied and at this stage they are purely speculative. Perhaps in prokaryotes the initial transcript of pri-miRNA may behave as mature miRNA, and thus in collaboration with the RISC complex may block the translation of mRNA either to temporarily halt the excess growth of the bacteria to prevent from exhaustion (or burn out), or to slow down the growth protein synthesis in an attempt to deliberately slow the growth of the bacteria. This is done in order to protect it from foreign viral multiplication in the cell or to totally or partially degrade the viral mRNA to protect the bacterial cell as part of its defense mechanism. Perhaps pri-miRNA may be produced or transcribed as a single stranded molecule as opposed to double-stranded in the eukaryotic cells. Since prokaryotic miRNA (memiRNA) are significantly smaller than mRNA, they may move out of the bacterial cell by the aid of a membrane transport system assisted by permease enzymes. Earlier in our laboratory, using radioisotopes as selective substrate markers, we proved that the entry of the nutrients or exit of the intracellularly produced metabolic end products in the probiotic bacterium Streptococcus thermophilus was controlled by the membrane transport system rather than through physical diffusion. If the miRNA of prokaryotic cells (memiRNA) can move out of the cells, along with other intracellularly produced peptides, organic acids and bacteriocins etc., the growth end products, or immunomodulins, must have memiRNAs as one of the essential integral constituents of the probiotic immunomodulins. If such memiRNAs of probiotics do exist in the immunomodulins, the question here is what is their role in maintaining the homeostasis of the Microbiota? In this context, microbiota is the total number of microorganisms present in the human GI tract. There are over 100 trillion bacteria represented by over 1000 species and genera in GI tract. Microbial cells are 10 times more than the total number of eukaryotic cells in the human body. The total number of genes in all the microorganisms present in microbiota, is called the microbiome. In essence, there are more microbial genes in the human body than human genes. To the best of my knowledge, no such investigations have been done before. This research project is undertaken to study, understand, and hypothesize the role of Prokaryotic miRNAs in inhibiting pathogenic microorganisms as well as their influence on the production and subsequent interaction with the eukaryotic miRNAs to inhibit the pathogens or modulate to maintain the optimal composition of the Microbiota and Microbiome to protect or improve overall human health. Since probiotics and their immunomodulins may be involved in coherence with eukaryotic immune cells to modulate and maintain the homeostasis of human Microbiota, it is worthwhile to review their mechanisms in order to understand the additional role of the miRNAs in this scenario. Now let us look into how probiotics recognize the pathogens in the human system.
Biochemical mechanism of prokaryotic probiotics to modulate the immune response in the Gastrointestinal tract:
Therapeutic probiotic bacteria recognize pathogenic bacteria by interacting with specific molecular patterns on the pathogen surface, known as “Pathogen Associated Molecular Patterns (PAMPs) with the aid of receptors on the probiotic cells called “Pattern Recognition Receptors (PRRs)” which allow them to distinguish between beneficial non-harmful bacteria from harmful pathogenic bacteria. After recognition of the pathogens, probiotic bacteria trigger immune responses to combat said pathogens. This is essential for maintaining gut health through prevention of pathogenic microbial infections. The PRRs present on the beneficial probiotic cell walls will bind to the specific PAMPs found on pathogenic bacteria. In the case of Gram-Positive pathogenic bacteria PAMPs are associated with lipoproteins, whereas as in the Gram-Negative pathogens they are associated with lipopolysaccharides. It is of interest to note that a major type of PRRs are Toll-Like Receptors (TLRs), which play a significant role in recognition of pathogens. There are several different TLRs in probiotic bacteria which can recognize several PAMPs of the variety of pathogenic bacteria. Once the probiotic PRR binds to the PAMP of the pathogen, it initiates a signaling cascade within the probiotic cell, leading to the production of several Antimicrobial substances and/or immune modulating molecules that can inhibit pathogenic microbial growth and/or activate the human immune system. In addition to this phenomenon, probiotics also compete for nutrients and attachment sites in the intestinal mucosa to prevent the colonization of pathogenic bacteria. Also, probiotics by interacting with the human immune cells, stimulate and activate the host immune response to combat the pathogenic microorganisms, perhaps by stimulating the production of specific eukaryotic miRNAs, which can also inhibit pathogenic bacteria in association with prokaryotic immunomodulins including memiRNAs. Having established these systems, let us look into how the human immune system recognizes pathogens.
Biochemical mechanism of human innate immune cells to recognize the pathogenic microorganisms and to activate cytokine and interferon production along with miRNAs:
In addition to probiotics, the cells involved in the innate immunity, i.e. Macrophages, Dendritic Cells (DCs) and Intestinal Epithelial Cells (IECs) can also initiate the immune response by recognizing MMPs thru PRRs since TLRs present in PRRs can detect molecules of bacterial, viral, and fungal origin. Just like probiotics, these innate immune cells will also have PRRs on the surface. After the TLRs of immune cells recognize a ligand of the pathogenic bacteria, they initiate an intracellularly signaling cascade, wherein the response proteins are activated or blocked, thereby activating the transcription factors that change the expression of several immune response genes, thereby releasing molecular mediators such as Interferons and Cytokines. Thus the orchestrated response activates various signaling pathways (thru alteration of the gene expressions) essential for immune defense mechanism, involving both inflammatory and anti-inflammatory processes. In addition, eukaryotic miRNAs also get involved significantly in this process. These miRNAs play a crucial role in orchestrating the immune response by regulating the gene expression at the post transcriptional level. Thus, miRNAs help fine tune the immune response, ensuring a balanced and effective defense mechanism. It has also been suggested briefly in literature that miRNAs produced by eukaryotes may act as modulators of molecular mediators in concert with the probiotic bacteria to kill pathogens and restore human gut health etc. If you look at the involvement of probiotics and the innate immune relating cells, it is obvious that their activities are very synergistic and interdependent. Yet such a relationship has not been studied thoroughly because there is not much research conducted on prokaryotic miRNAs (memiRNAs) and their interactions with eukaryotic miRNA.
The current investigation is undertaken to determine if probiotic bacteria produce any miRNA – like memiRNAs – to control the physiology of the cell under normal conditions and also under distressed conditions such as an infection by bacteriophage.
Additionally, the goal is to check if such memiRNAs are in fact part and parcel of the probiotic-produced immunomodulins. Now one can understand the role of microRNAs in the eukaryotic cell; the majority of these microRNAs also prevent transcription of several genes into (miRNA). Currently this investigation is undertaken to study the role of miRNAs in the prokaryotic cell. Particularly my attention is on therapeutic probiotic bacteria. Do probiotics produce microRNAs (memiRNAs), and if so, what are their functions in the regulation of modulating the Microbiota? Limited research in this arena has proved that microRNAs in prokaryotic cells (memiRNAs) are not quite micro because unlike eukaryotic miRNAs, prokaryotic ones are larger in size, being close to 100-500 nucleotides long. Yet they are quite small compared to the number of nucleotides present in mRNAs of prokaryotes. The larger size of the microRNAs of the prokaryotic cells (memiRNAs) in comparison to eukaryotes (miRNAs) are perhaps due to the lack of a nuclear envelop in bacteria, and thus to start with they may be transcribed as a single-strand mature microRNAs without going thru further modification, as in the case eukaryotes. The relatively smaller size of the memiRNAs, compared mRNA, etc. facilitates their exit from the bacterial cell in the form of vesicular microRNAs along with other intracellularly produced bacterial growth end products. Perhaps vesicular memiRNAs might exit from the bacterial cell through a specialized membrane transport system, mediated by permease enzymes. As I have indicated earlier, since probiotic-associated microRNAs are larger than the eukaryotic cell microRNAs (miRNAs), for the first time I would like to introduce a new term called mega-microRNAs, abbreviated as memiRNAs to distinguish them from eukaryotic miRNAs. Hereafter in the remainder of this text, prokaryotic microRNAs will be referred to as memiRNAs, and eukaryotic microRNAs as miRNAs.
What exactly is the function of these memiRNAs in probiotic bacterial cells?
This is where our curiosity started to find out more about these memiRNAs. While working on the novel discovery/ invention involving multiple mixed strain probiotics along with their immunomodulins and their positive effect on preventing and/or curing the SARS-CoV-2 coronaviral infection, we discovered that probiotic-produced immunomodulins are significantly more effective in inhibiting a Corona viral Covid-19 infection than the probiotic bacteria. We have attributed this to specific short chain fatty acids, bacteriocins, hydrogen peroxide, and organic acids produced by probiotic bacteria. We have also mentioned the term Non-Specific Inhibitory Compounds with reference to the composition of the probiotic produced immunomodulins since we thought in all probability, there could be some other functional factor besides the normally known inhibitors. My question here is: if such additional compounds were to be present in immunomodulins, would they be the probiotic-produced microRNAs, i.e. memiRNAs? We have further investigated to study and elucidate this unknown phenomenon. Although the novel patent has been issued (US Patent # 11,077,052 B1) the exact single and crucial therapeutic compound that was responsible for the prevention or suppression of the SARS-CoV-2 virus causing Covid-19 infection and other secondary bacterial infections was not fully investigated. Thus the following experiments were conducted to elucidate the specific or multiple therapeutic factors present in probiotic-produced immunomodulins. One such curiosity factor was the obscure reason(s) for probiotic bacteria exhibiting no further growth once they attain their maximum growth (stationary phase) no matter what other alterations were made such as neutralizing acidic pH, supplying fresh nutrients, detoxifying hydrogen peroxide in the growth medium, etc. Yet probiotic bacteria will not grow any further and still however maintain their viability for up to a certain length of time depending on their genetic makeup, which varies from strain to strain. I have suspected that this stoppage of growth can be due to involvement of probiotic microRNAs (memiRNAs) to stop the translation of the growth proteins.
In addition, at this juncture we thought the lack of further growth may be due to the lack of space in the micro-environment to compact more bacteria, or perhaps maybe due to contact inhibition.
Even if it is in fact due to contact inhibition, it can only be attributed to the involvement of some kind of genetic variation within the cell. However, to verify these factors, we centrifuged fully-grown probiotic cultures at high speed to see if we can further pack the cells. Here through centrifugation, we were able to compact them over tenfold, indicating it was not a space issue. Then what is stopping their growth at their stationary phase? What is happening in the stationary phase besides the adverse conditions in the growth medium, such as nutrient depletion, end product inhibition, adverse acidic pH, presence of excess organic acids and hydrogen peroxide etc.? Although we have corrected all these adverse inhibitory factors and supplied them with fresh nutrients, still the probiotic bacteria did not multiply. Yet, when we transferred a small amount of such cell mass into fresh medium, they multiplied at a fast pace, indicating that the growth cessation was only temporary. We wanted to investigate the factor(s) that temporarily controlled or inhibited growth. At this stage we started to suspect that growth cessation can be due to prevention of translation of the essential growth proteins due to blockage of mRNA expression through binding of memiRNA. Perhaps such memiRNAs can also be vesiculated and excreted into the extracellular growth medium along with other growth end products, which are collectively called immunomodulins. If so, what is the role of this memiRNA in inhibiting viral multiplication, which we have observed in the treatment of Covid-19 using RT-PCR test, as outlined in the novel US patent # 11,077,052 B1?
In this connection it is worthwhile to mention that another US patent has also been issued in 2023 (US patent # 11,643,641 B2) with reference to the role of naked corona viral RNA in inducing Covid-19 infections, and measures taken to inactivate this naked RNA (viral genome) in order to curb the pandemic.
Viral naked RNA should not be confused with microRNA or memiRNA. It is all pointing out the importance of RNAs and microRNAs to cause or combat viral pandemics, cancers, nosocomial infections, and autoimmune diseases. United Stated patent (US patent # 6,080,401) outlined a novel invention of using plant-based herbs as therapeutic ingredients along with probiotics, which significantly improved the drug efficiency of the herbs to cure diseases faster and without any side effects. Since it has been pointed out later that microRNAs also exist in plant-based products, we thought the success of the patented invention to curb diseases perhaps was due to the combination of plant based as well as the probiotic produced microRNAs (memiRNAs) in conjunction with the therapeutic principles of the herbs and the probiotic immunomodulins, along with the eukaryotic produced microRNAs under the influence of such therapies. It may be entirely possible that part of the therapeutic effect of the herbs could also be due to plant-based microRNAs. According to Reddy, immunomodulins produced by probiotics have exhibited superior antiviral and antibacterial properties compared to their parental probiotics, although both are essential. Thus this investigation is undertaken to study the unknown factors in probiotic-produced immunomodulins, specifically the role of memiRNAs.
In order to obtain answers to several of these unknown factors, the following schematic and systematic experiments were conducted on probiotic bacteria and their end products/ immunomodulins, produced under various conditions, which are outlined in the following materials and methods section.
Materials and Methods
The following experiments (1 through 5) were conducted systematically:
- To determine the maximum probiotic bacterial cell numbers produced in a growth medium using an external pH control system, and further centrifugation of such cell mass to compact them in an attempt to determine if cell growth stoppage was either due to limitation of space in the micro-environment of the growth medium or due to some other factor such as intracellular microRNAs (memiRNAs) blocking the translation of the growth proteins.
- To determine the possible factors to improve the physiological activity of probiotic bacteria after they attain the maximum cell growth, and after they are further concentrated through centrifugation, to check for the involvement of probiotic produced microRNAs (memiRNAs).
- The effect of probiotic growth spent medium (separated from the bacterial cells), which is termed immunomodulins, on inhibiting the host probiotic bacterium as well as E.coli bacterium, to assess inhibitory patterns.
- Determination of the principal causative factor in the immunomodulins produced by Streptococcus thermophilus to inhibit E.coli, besides the known established inhibitors, specifically to look for RNA fractions I.e. microRNAs (memiRNAs).
- Determination of the inhibitory factors present in the phage lysate, specifically to check for probiotic-produced microRNA i.e. memiRNAs, (produced under the influence of bacteriophage) using the host bacterium Streptococcus thermophilus. Also to investigate the effect of phage-induced lysis of one probiotic strain on the growth and activity of other probiotic strain(s) belonging to different genera, specifically in terms of memiRNAs involvement in retardation or activation of specific cell growth proteins. These experiments were designed to study the influence of bacteriophages on induction of the microRNAs of probiotic bacteria (memiRNAs), under stressful conditions, which are generally encountered in the GI tract microbiota. In the GI tract microbiota, limited research proved that naked RNA of the coronavirus can penetrate into the bacterial members of the human microbiota-more like phage.
Experiment 1:
Streptococcus thermophilus strain ST- 2 was inoculated into 100 gallons of the sterilized growth medium and incubated at 32 C. The growth medium was formulated using the following ingredients: Sweet whey solids (50 %), Nonfat dry milk (20 %), Calcium-phosphate buffers (10 %), Yeast Extract (7.5%), Glucose (10 %), various major and minor minerals (2.5 %). The medium was reconstituted to 15 % solids, prior to sterilization. The automatic pH control system (pH probe) using ammonium hydroxide as a neutralizer was attached to the growth vessel to neutralize the growth medium, as culture grows, to maintain pH between 5.1 to 6.0, to eliminate the growth inhibition due to adverse low pH. Since it is on the external pH control system, as the probiotic bacteria grow and produce lactic acid and other end products, the neutralizer ammonium hydroxide was continuously discharged into the growth medium to maintain the pH between 5.1-6.0. Roughly after 16 hours the growth stopped indicated by the stoppage of the neutralizer being introduced, observed by the automatic pH register. At this stage, the fully grown culture was examined under microscope, activity test was conducted using Ellikers activity test. The Ellikers activity test conducted was as follows: The culture was inoculated at 3% rate into 10 ml. of sterilized 10% solids reconstituted antibiotic free Non-Fat Dry Milk and incubated at 37.5 C for 3.5 hours. The entire contents were titrated with 0.1 normal NaOH to a faint pink color using phenolphthalein as an indicator. The results were recorded as percent Lactic Acid. In addition, The fully grown culture was plated to determine the total bacterial numbers using the standard Enriched Tryptic Soy Agar. At this stage although a fresh sterilized nutrient was introduced, the multiplication of the probiotic bacteria stopped, indicating that the growth phase ended, and it came to a halt. To improve the cell numbers, the fully grown culture was centrifuged, and the cell mass was collected, discarding the growth end products. The concentrated cell mass was examined under microscope, and the total bacterial numbers were determined by plating using the enriched tryptic soy agar. The Ellikers activity test was also performed. The results of this experiment are presented in Table-1. It was observed that when once the probiotic bacteria grow to attain a maximum cell number, using external pH control system, it was next to impossible to make them grow any further.
Experiment 2:
The experimental design and the growth conditions were like the first experiment. However, at this stage, besides the adjustment of pH towards alkalinity, filter-sterilized catalase enzyme was added to detoxify the hydrogen peroxide etc. The Ellikers activity test (as outlined in Experiment 1) was conducted after the cessation of growth and after centrifugation (concentrated cells). The activity test results are presented in the results and discussion section (Table-1). Although the growth of probiotic bacteria stopped at certain stage, they have exhibited excellent growth after they were transferred and grown in a fresh nutrient medium (reconstituted Non-Fat Dry Milk). More activity was observed with the centrifuged cell mass. At this stage we suspected that there is some other factor besides the adverse pH, low level of nutrient, and inhibitory factors such as hydrogen peroxide etc. which was inhibiting the further growth of probiotic bacteria. The following experiment was conducted to check for the possible additional reasons for the cessation of further growth of probiotic bacteria.
Experiment 3:
The experimental design for growing the probiotic bacteria was same as the Experiment 1. However, at the end of the growth phase, the grown culture was centrifuged and therefore separated the bacterial cells from the spent broth. The spent broth was designated as the immunomodulin fraction. This was subjected to Millipore filtration using a 0.22 milli micron filter to totally remove any traces of the bacteria. Prior to filtration, the pH was adjusted to 7.0 and catalase enzyme was added to remove any residual hydrogen peroxide. The filter sterilized preparation was tested to see if it could inhibit the parental probiotic bacteria and several other S. thermophilus probiotic bacterial strains and E.coli using disc assay. The results are presented Table 2. The results revealed that probiotic bacteria were not inhibited, yet the immunomodulins inhibited the gram negative E.coli bacteria, even though it was not due to low pH or due to the residual hydrogen peroxide etc. Thus, further experiments were designed to elucidate the causative factor(s) present in the probiotic produced immunomodulins, which caused E.coli inhibition.
Experiment 4:
The immunomodulins or the spent growth medium was collected as outlined in the previous experiment # 3. Probiotic spent growth medium (immunomodulins) were pH adjusted to pH 7.0, treated using catalase enzyme and then filter sterilized. Then it was divided into four fractions. Fraction 1, as is served as control; Fraction 2 was treated additionally with protease and amino peptidase enzymes to find out if these proteins or peptide fractions or any protein based bacteriocins were responsible for the inhibition of E.coli. Fraction 3 was treated with DNase enzyme to see if the inhibitory factor was associated with probiotic bacterial DNA or DNA fractions. Fraction 4 was treated with RNase enzyme, to check if the inhibitory factor was associated with the probiotic associated or produced RNA fractions. All the four samples, prepared as above, were tested on E.coli using Disc assay, to check for the inhibitions, to elucidate the causative factor of the inhibition. The results of this experiment are presented in Table 3.
Experiment 5
Streptococcus thermophilus ST-2 bacterium was inoculated into sterile 12 % solids reconstituted nonfat dry milk. It was divided into two fractions. Fraction 1 served as a control and fraction 2 was inoculated with the specific phage for S. thermophilus ST-2. These two fractions were incubated at 37 C for 12 hrs. At the end of the incubation both the total bacterial counts and phage counts were determined along with pH. Total bacterial counts were determined using the standard enriched Tryptic soy agar. The phage counts were determined using standard phage plaque assay. Then both the samples were neutralized to Ph 7.0 and treated with chloroform (0.5 ml chloroform to 15 ml. sample) to inactivate the S. thermophilus bacteria, but not its phage. After 10 minutes of holding the chloroform + sample, the samples were aerated by blowing sterile air to drive off the chloroform. Since S. thermophilus ST-2 bacteriophage tends to stick to millipede filters, chloroform was used to inactivate the host probiotic bacterium, but not the DNA phage. After 19 to 15 minutes chloroform was driven out by blowing sterile air, ending up with only viable bacteriophage. The samples were checked for the inhibition of probiotic S. thermophilus bacterium using disc assay. Since the S. thermophilus ST-2 phage was specific to its host, consequently inhibition was evident. To check to see if such inhibition exists in the phage unrelated Streptococcus thermophilus bacteria, several strains (6 others) were tested using the above sample 2. This is to check if there are any other inhibitory factors in the phage lysate, besides the ST-2 bacteriophage. Thus, the above phage lysate sample of the S. thermophilus was divided into four fractions as outlined in experiment 4 using a similar treatment. All the four samples were tested using disc assay on all other phage unrelated probiotic S. thermophilus bacteria. The results are presented under the results and discussion section (Table-4). This experiment was conducted to see if microRNAs produced and released under the influence of phage are different than the ones produced without phage infection. It is to be noted that naked coronaviral RNA can also penetrate the bacteria in microbiota-more like phage.
Results:
Results of Experiment 1:
Concentration of bacteria through continuous neutralization using the external Ph control resulted in the stoppage of neutralization after 16 hrs. of incubation, indicating that the probiotic bacterial growth stopped at this stage. Microscopy revealed that bacteria were packed, and they were not in chains, instead they were showing single cell morphology, which is an indication that cell growth ceased. The total bacterial count of the fully-grown culture was ten billion per milliliter. Although we have introduced more sterilized nutrients at this stage, the bacterial cell count did not improve. At this stage we thought it was due to limited space for the bacteria to pack in the vicinity or in the surrounding microenvironment of the growth medium. It was determined for sure that probiotic bacteria stopped multiplying. The general explanation given for such a stoppage of growth was that the culture has entered into the stationary phase or due to the end product inhibition or due to lack of additional nutrients or due to abnormally low Ph retarding the uptake of the nutrients due to cessation of the membrane transport system. Even though we have corrected all the above so-called inhibitory factors, yet the probiotic bacteria totally stopped multiplying indicating that there was some other factor in the cells which was stopping their further multiplication. When the fully-grown probiotic culture was centrifuged using high speed centrifugation, the cell mass was further packed and the total bacterial count was over 120 billion per ml, indicating that the lack of space in the microenvironment surrounding the fully-grown culture was not the limiting factor for the cessation of bacterial growth. Thus it was proven that the stoppage of growth was not due to adverse final Ph, lack of nutrients or limitation of space in the microenvironment surrounding the probiotic bacteria in the growth medium. The results of these experiments are presented in Table -1. This experiment proved that there is indeed some other factor which is inhibiting the further growth of probiotic bacteria, stemming within cells which have something to do with the bacterial genome.
Results of Experiment 2:
In this experiment, after the full growth of culture and further stoppage of additional growth, no matter which biochemical modifications were made, it did not result in alleviating the growth stoppage. Perhaps this growth cessation might have been due to the presence of probiotic produced biological hydrogen peroxide? Even after we treated the culture using the filter sterilized catalase enzyme to neutralize the probiotic-produced hydrogen peroxide, the culture still did not produce any additional cells. The total bacterial count was only 10 to 12 billion organisms per ml. Also, the activity test conducted before and after the catalase treatment was same indicating the growth cessation or stoppage was not due to integral probiotic produced hydrogen peroxide. The results are presented in Table-1.
| Hrs. of Incubation Prior to Centrifugation | Bacterial Counts/ ml Prior to Centrifugation | Activity Test Results Prior to Centrifugation | Bacterial Counts and Activity of Centrifuged Mass |
|---|---|---|---|
| 12H | 70 x 107 | 0.37 Bacterial Count Activity | |
| 14H | 150 x 107 | 0.4 | |
| 16H | 100 x 108 | 0.6 | 120 x 109 0.75 |
| 18H | 110 x 108 | 0.58 |
The fully-grown probiotic culture treated with the catalase enzyme and then centrifuged to concentrate the cells had a total count of 120 billion per ml or gram, which was about 10 fold increase due to concentration. Also, the activity test was significantly higher than the not concentrated (before centrifugation) probiotic culture. At this stage it was confirmed that the probiotic bacterial growth stops at one stage due to some other molecular factor other than the lack of nutrients, adverse pH, and inhibition due to molecular hydrogen peroxide produced by the probiotic bacteria etc. It was suspected at this stage that perhaps the auto inhibition of the probiotic bacterial growth can be due to some of the therapeutic peptides or due to some of the DNA or RNA related fragments which were excreted off the probiotic cells or some other microRNA type elements produced within the probiotic cells which were blocking the translation of the essential proteins required for the cell multiplication, as observed in the eukaryotic cells.
Results of Experiment 3:
The fully-grown Streptococcus thermophilus culture was centrifuged and the soluble growth end products or immunomodulins were filter sterilized and catalase treated. It was checked to see if they can inhibit the growth of the parental (the bacteria which produced the immunomodulins) probiotic bacteria, as well as the E.coli using the disc assay. The test results proved that the parental bacteria (S. thermophilus ST-2), which produced such immunomodulins was not inhibited by them. Also, several other strains of S. thermophilus bacteria tested were not inhibited. However, the probiotic produced immunomodulins or growth end products exhibited significant inhibition of the E.coli, indicating that the S. thermophilus produced immunomodulins were not the causative factors to induce the cessation of the probiotic bacterial growth in the fermenter. The results of this experiment are presented in Table-2. Once again it is proven that there is some other molecular mechanism operating within the probiotic bacterial cell causing the prevention of the multiplication of the probiotic bacterial growth after they attain their full growth.
| Inhibition Pattern of Streptococcus thermophilus ST-2 Immunomodulins On: | Presence or Absence of Inhibition |
|---|---|
| Streptococcus thermophilus ST-2 | Negative |
| Streptococcus thermophilus ST-4 | Negative |
| Streptococcus thermophilus ST-A | Negative |
| Streptococcus thermophilus ST-C | Negative |
| Streptococcus thermophilus ST-G | Negative |
| Streptococcus thermophilus ST-X | Negative |
| E.coli | Positive |
Results of Experiment 4:
Fraction 1, which is the spent broth of the probiotic bacterial growth where the Ph was adjusted to 7.0 and hydrogen peroxide has been neutralized using catalase enzyme exhibited significant inhibition of E.coli (++++). Whereas fraction 2, where in, in addition to the above treatment, protein and peptide fractions (including bacteriocins) were destroyed using the protease and aminopeptidase enzymes, still exhibited significant inhibition of E.coli (+++). It has been proven that that inhibition was not due to either therapeutic proteins or peptides, including bacteriocins of the probiotic produced immunomodulins. Fraction 3, where in the treatment was same as in fraction 1, except additionally it was treated with the DNase enzyme. This fraction also inhibited E.coli same as fraction 1 and 2, indicating that the inhibition was not due to DNA associated fragments of the probiotic bacteria. Fraction 4, where in the treatment was also same as fraction 3, except instead of DNase enzyme RNase was used. Surprisingly fraction 4, did not inhibit the E.coli, indicating that the inhibitory factor perhaps could be an RNA fragment belonging to the parent probiotic bacteria, which was excreted or membrane transported from the probiotic bacterial cells into the immunomodulins fraction, towards the end of the growth phase. Apparently, In order to come out or transported out of the bacterial cells as a vesicular RNA, it has to be small in size, (although we have not determined it) perhaps it could be in the tune of 100 to 200 nucleotides length. Earlier it was proven that the procaryotic microRNAs (even though they were not called microRNAs) were around 100 to 500 nucleotides length, as opposed to eukaryotic microRNA which is around 20 to 25 nucleotides length. It was proven beyond doubt that the inhibitory factor was excreted from the probiotic cells towards the end of their growth period, and it was not inhibitory to the probiotic bacterial growth which produced it, and yet it was inhibitory to E.coli. The results of this experiment are presented in Table 3.
| Number | Variable | Inhibition Pattern on E.coli |
|---|---|---|
| 1 | Spent Broth – pH Adjusted 7.0 – Catalase Treated | ++++ |
| 2 | Spent Broth – pH Adjusted 7.0 – Catalase Treated- Protease and Amino Peptidase Treated | +++ |
| 3 | Spent Broth – pH Adjusted 7.0 – Catalase – DNase Enzyme Treated | +++ |
| 4 | Spent Broth – pH Adjusted 7.0 – Catalase – RNase Enzyme Treated | Negative |
| 5 | Spent Broth – pH Adjusted 7.0. – Catalase – RNase added first and at end Protease-Amino Peptidase Added | Negative |
| 6 | Catalase Enzyme by Itself | Negative |
| 7 | Protease-Amino Peptidase by Itself | Negative |
| 8 | DNase by Itself | Negative |
| 9 | RNase by Itself | Negative |
Although we did not test on other genera of pathogenic bacteria, in all probability it may also be inhibitory to them. At this stage we were to investigate if such probiotic microRNA can be induced as part of the defense mechanism by probiotic bacteria, when challenged using their bacteriophage. Also, to find out if they exhibit similar inhibitory patterns as the ones produced without phage infection.
Results of Experiment 5:
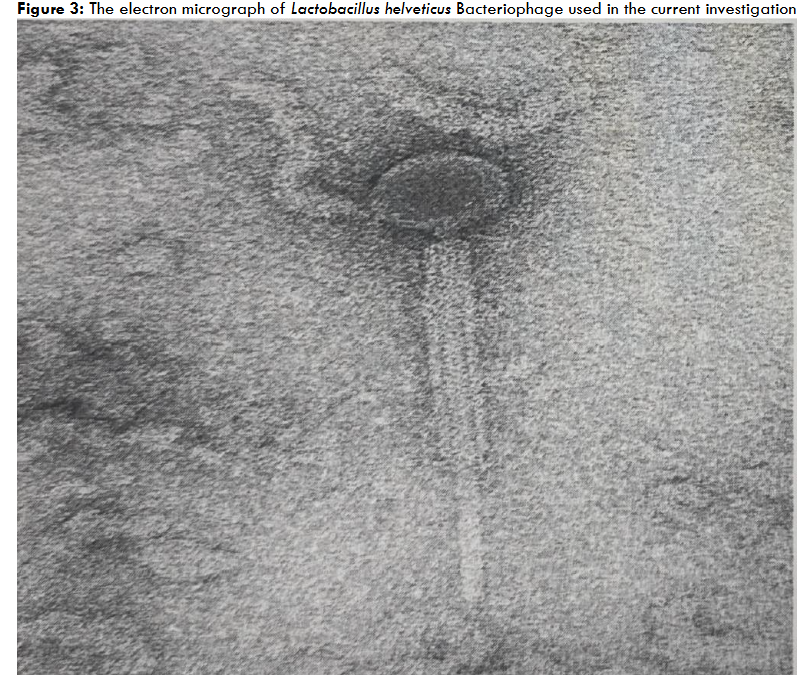
The Streptococcus thermophilus ST-2 phage lysate exhibited inhibition of the several phage unrelated-yet belonging to same genera and species probiotic strains of Streptococcus thermophilus. Phage plaque assay proved that the inhibition was not due to phage, since ST-2 phage was only specific to inhibit S. thermophilus probiotic bacterial strain ST-2 only. Electron microscopy also revealed the presence and morphology of the S. thermophilus bacteriophage as shown in Figure 1 and 2. Electron Microscopy was conducted to verify and confirm that the specific ST-2 bacteriophage induced lysis than the morphologically different other bacteriophages belonging to other species.


At this stage, we thought the inhibitor could be phage lysate relating to the peptidoglycan breaking enzymes induced by the phage as part of the lytic cycle. We were proven wrong because protease enzyme treatment did not block inhibition, proving that it was not due to lytic enzymes. Out of all the four fractions, whose preparation has been outlined under the material methods section of this article, the fraction 4 treated with RNase enzyme did not inhibit all the strains of S. thermophilus included in the study, other than the S. thermophilus ST-2 strain, which was inhibited due to its own specific phage. It was evident from this experiment that a phage induced microRNA produced by S. thermophilus ST-2 was the culprit for inhibiting the gram-positive closely related S. thermophilus strains. The results are presented in Table-5. Perhaps different microRNA, which had a totally different function in the intact host cells, ended up in the phage lysate due to cellular lysis by bacteriophage, thus inhibiting multiple probiotic strains of the S. thermophilus. Apparently, the microRNA of the procaryotic probiotic bacterium (memiRNA) was produced in the bacterial cell as an aid to suppress the growth rate of the bacterium in an attempt to override or slow down the multiplication of phage or spread of the phage particles in an attempt to protect the host bacterium. This explains why some of the mutants of the S. thermophilus turn into phage resistant cultures, after most of the bacteria are lysed by the virulent phage. This particular serendipitous observation will have tremendous application in terms of using bacteriophage to induce probiotic microRNAs (memiRNAs), which can be used as therapeutic agents to control the pathogenic infections, including the multiple antibiotic resistant nosocomial bacterial infections, such as MRSA and C.diff etc.
| Number | Variable | Inhibition Pattern on E.coli |
|---|---|---|
| 1 | Ø Lysate – pH Adjusted 7.0 – Catalase Treated | ++++ |
| 2 | Ø Lysate – pH Adjusted 7.0 – Catalase Treated-Protease and Amino Peptidase Treated | +++ |
| 3 | Ø Lysate – pH Adjusted 7.0 – Catalase – DNase Enzyme Treated | +++ |
| 4 | Ø Lysate – pH Adjusted 7.0 – Catalase – RNase Enzyme Treated | Negative |
| 5 | Ø Lysate – pH Adjusted 7.0. – Catalase – RNase and at end Protease-Amino Peptidase Added | Negative |
| 6 | Catalase Enzyme by Itself | Negative |
| 7 | Protease-Amino Peptidase by Itself | Negative |
| 8 | DNase by Itself | Negative |
| 9 | RNase by Itself | Negative |
These experiments (Experiment 1 and 2) proved that probiotic bacterial growth cessation, at the end of the growth phase, can be due to microRNA produced intracellularly to halt the production of growth promoting proteins by binding to the mRNA coding for such protein synthesis. Perhaps it explains why in nature such a mechanism exists so that bacteria cannot keep on multiplying to such an extent to take over the world and disturb the Eco Balance. To the best of our knowledge, this is the first investigation to elucidate the mechanism of auto inhibition of probiotic growth due to microRNA produced within the bacterial cell. This investigation has also proved that probiotic produced microRNA either in the form of vesicular or non-vesicular will end up in the extracellular growth end products thru excretion or through permease mediated transport system from the probiotic bacterial cells, at certain stage of the bacterial growth cycle.
Although we did not determine the number of nucleotides in the probiotic produced microRNA, according to the limited research done by other investigators, it should be in the vicinity of 100 to 500 nucleotides length, which is much bigger than eukaryotic microRNA, yet behaves and functions like the eukaryotic microRNA. Thus, I would like to give a new nomenclature or name it as mega-microRNA or in short memiRNA. In addition, this investigation also proved that bacteriophage can be used to induce the production of micro RNA (memiRNA) from the procaryotic bacterial cells, which can be used as therapeutic pharmacological ingredients to inhibit or treat the pathogenic bacteria, including antibiotic resistant hospital associated or nosocomial infections, which are killing close to million innocent people in the world. According to the scientific projection, by the leading scientists around the world, by the year 2050, the annual death rate due to this hospital associated infections will far exceed over 50 million, which will be significantly much bigger than the annual cancer deaths of 10 million.
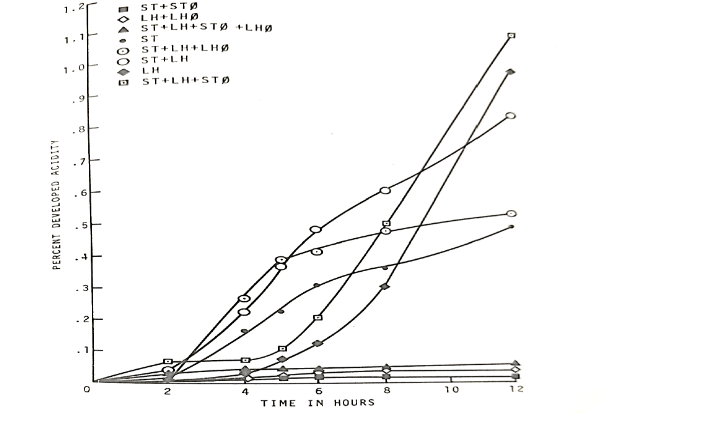
The second experiment which is a part of the experiment 5, using the Streptococcus thermophilus ST-2 bacteriophage to lyse the S. thermophilus when it was growing as mixed probiotic culture in combination with the Lactobacillus helveticus and vice versa, revealed the following unexpected results which are presented in table 6 and figure 5.
The percent developed acidity at the end of 12 hrs. incubation when both the S. thermophilus and L. helveticus were grown together was 0.80. Whereas S. thermophilus and L. helveticus when grown separately exhibited the percent developed acidities of 0.45 and 0.95 correspondingly. When L. helveticus phage was inoculated and grown along with S. thermophilus and L. helveticus culture, the percent developed acidity was 0.50. However, when S. thermophilus phage was grown along with S. thermophilus and L. helveticus, the percent developed acidity was 1.15. These results prove that the phage lysate of the S. thermophilus had a significant stimulatory effect on L. helveticus bacterium. Similar results were obtained when L. helveticus was replaced by other strains belonging to L. lactis and L. bulgaricus.
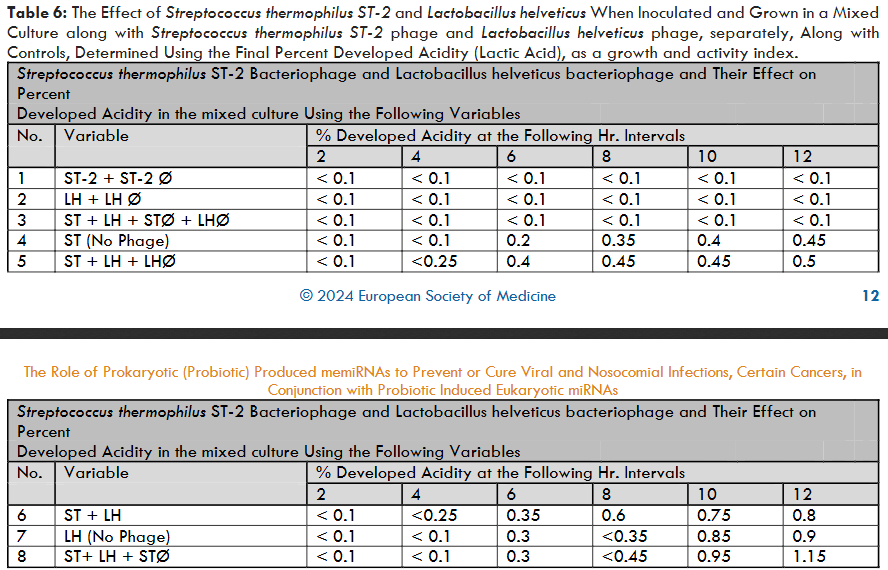
This is an unexpected result wherein apparently the microRNA produced by the probiotic S. thermophilus under the influence of its bacteriophage enhanced the growth of the probiotics L. helveticus, L. lactis, and L. bulgaricus. It goes to prove that the S. thermophilus phage lysate must have some microRNAs, which were produced perhaps to stop or slow down the mRNA coding for the specific growth proteins by blocking the translation, in order to protect the host. Such a microRNA produced under the influence of bacteriophage of S. thermophilus, might have blocked the transcription of specific microRNA of the probiotic L. helveticus by blocking the gene coding for such microRNA, which was intended to block the growth of L. helveticus after it attains its maximum growth.
Similar stimulation of S. thermophilus was also observed when L. helveticus phage was inoculated and grown together with S. thermophilus and L. helveticus. However, it was not as significant or obvious as we have observed with S. thermophilus lysis in the mixture of S. thermophilus and L. helveticus. Additionally, electron microscopy was also conducted on the samples to verify the presence of L. helveticus phage. The electron micrographs of L. helveticus phage are presented in Figures 3 and 4. It goes to prove that the microbial probiotic microRNAs can serve both as inhibitors on some probiotics and growth stimulators on other probiotics.
It explains the role of prokaryotic probiotic microRNA to stabilize or harmonize the human gut Microbiota and Microbiome by balancing the bacterial species to assist the immunomodulation to stimulate or simmer the immune systems depending on the physiological condition of the human subject. Also, such immune modulation exerted by the microRNA of the probiotics, along with the probiotics by themselves, may also assist the production of human eukaryotic micro RNA by the human epithelial cells to produce and excrete into the gastrointestinal tract as fecal microRNA, to inhibit the unwanted pathogenic bacteria, including the antibiotic resistant nosocomial infection causing microbes.
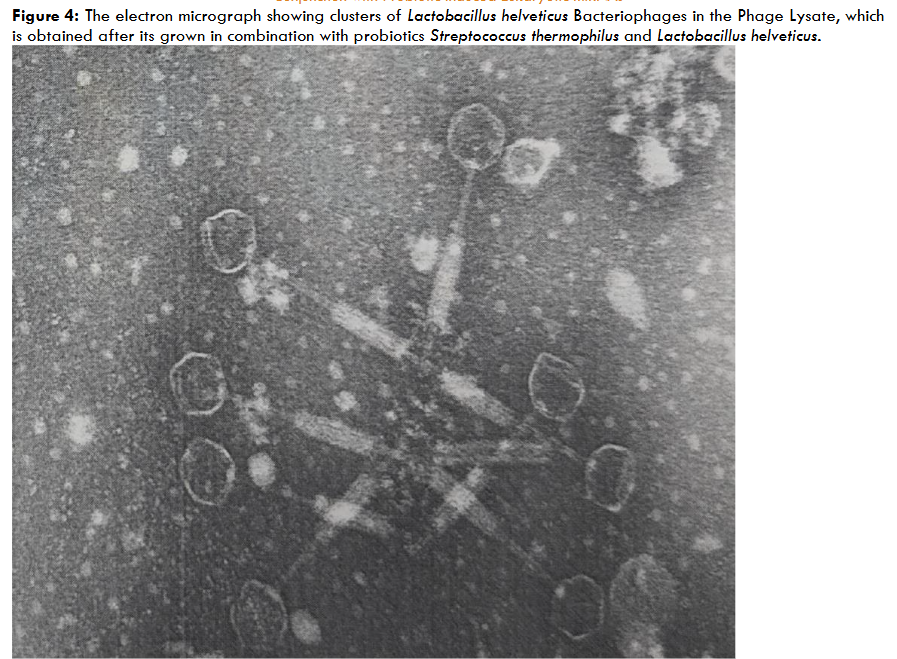
Once again, these experimental findings proved that the bacteriophages of the probiotics may have significant effect on production and modulation of microRNAs to harmonize and balance the human gut Microbiota and Microbiome. Although such systems could have been operating in the healthy human beings naturally, these observations further assist to develop specific phage therapies by using phage as a mediator to stimulate the production of the prokaryotic microRNAs which can be used as a therapeutic pharmaceuticals rather than using phage itself directly as a therapeutic ingredient to override the limitations imposed by the immune system.
Figure 5: Effect of cellular lysis by bacteriophages (Ø) on associative growth of Streptococcus thermophilus ST-2 (ST) and Lactobacillus helveticus (LH) strains as measured by acid production.
Discussion:
The results of these experiments are pointing our thinking in a different direction. For time immemorial, microbiologists have always been under the impression that the growth phase of bacteria ends at the stationary phase due to nutrient depletion or growth end products inhibitions or adverse low pH interfering with the absorption of the nutrients or some unknown genetic factor etc. No previous investigator looked into the possibility of the cessation of the growth at the stationary phase being due to the non-coding microRNA blocking the translation of the essential growth promoting proteins. This investigation proved that microRNA is indeed one of the factors to limiting the growth of bacteria in order to protect them from dying, going into a stage of burn out or mutation resulting in weaker progeny once they grow to their maximum limit. In addition, strain dominance among bacteria when they are grown together with other bacteria was also attributed earlier to the inhibition exerted by one bacterium over the other, either due to competition for nutrients or the production of inhibitory factors by the dominant bacteria over the recessive strains etc. This investigation cast a different light on this subject by bringing microRNAs role into microbial strain domination, which is as follows: Earlier, the breakthrough invention of Reddy et al., differential agar(s) and differential broth to identify and differentiate the closely related species of the lactic streptococci on a single agar medium and single broth, for the first time in the world, opened up a new science and methodologies for studying the associative growth relationships among the closely related bacteria when they are grown together using a direct differential agar, which differentiates three species on the same agar plate without having to go through studying individual colonies to identify the strains.
The published results of Reddy et.al.’s investigation in the late 1960’s and early 1970’s proved that some strains of probiotic lactic streptococci such as Streptococcus diacetylactis dominated by suppressing the closely related strains belonging to Streptococcus lactis and Streptococcus cremoris. The authors in 1970’s attributed this strain dominance to nutritional competency and other unknown inhibitory factors such as rate of growth, byproduct inhibition, etc. The results of my current investigation proves that there is another significant factor which can explain the mechanism behind strain dominance among closely related probiotics proven by the fact that different bacterial strains attain stationary phase at different times due to memiRNAs inhibiting the growth proteins, which in turn depends on the inherent genetic characters of each strain etc. The cessation of bacterial growth perhaps is controlled by the microRNAs (memiRNAs) produced within the cells to prevent or retard the translation of specific growth promoting proteins by binding to their mRNAs. Thus, S. lactis culture may produce intracellularly a specific microRNA (memiRNA), which can halt its growth perhaps as early as 12 hrs. of incubation. Whereas S. cremoris may attain cessation of growth hypothetically around 14 hrs., indicating the onset of production of microRNAs (memiRNA) may be slightly delayed than in the case of S. lactis. In the case of S. diacetylactis the role of microRNAs (memiRNAs) may be slightly less or perhaps the production and action of its intracellular memiRNAs are significantly delayed in comparison to its closely related, yet different strains of S. lactis and S. cremoris.
Contrary to the thinking of traditional microbiologists, perhaps the role and activity of microRNAs (memiRNAs) dictate the organism’s ability to dominate when grown together with other microorganisms. Such a phenomenon of strain dominance may be going on a routine basis in the gastrointestinal tract among members of the microbiota, resulting in either good or bad associative growth relationships among bacteria, and resulting in either negative (dysbiosis) or positive variance in the microbial composition of the Microbiota. This may have excellent application on developing multiple mixed strain probiotics, which will grow without exhibiting any strain dominance in the Gastrointestinal tract to maintain homeostasis and the optimal composition of healthy Microbiota and Microbiome to ensure good health and immunity to the host. As of today, nobody is taking advantage of using procaryotic microRNAs for the benefit of humanity, although probiotics were granted GRAS status by FDA. Part of the reason is perhaps such research is at infancy and not too many researchers are involved in this arena, especially in the field of probiotics.
United States Patent # 11,077,052 B1 was issued after severe scrutiny regarding the role of multiple mixed strain probiotics along with their immunomodulins in preventing or treating COVID-19 infection due to SARS- CoV-2 RNA virus. Although partial explanation was given regarding physiology behind the success and validity of the novel invention, no mention was made regarding the role of probiotic produced microRNAs and even their presence in the probiotic produced immunomodulins. Perhaps, the scientific community never considered such functional microRNA existed and operated in procaryotic micro-organisms, specifically in probiotics. However, the patent clearly stated regarding the presence of organic acids, bacteriocins, and therapeutic peptides etc., present in the probiotic produced immunomodulins, are the key viable factors to inhibit the SARS-CoV-2 Coronavirus. In addition, the patent also stated that immunomodulins must have some other non-specific inhibitory compound(s) which may be playing a significant role in inhibiting coronavirus multiplication, and other Covid-19 associated secondary pathogenic microbial infections. The invention outlined in this publication partially clarified that such non-specific inhibitory factor may be microRNA produced by the probiotic bacteria. Earlier, it has also been stated in US Patent # 11,077,052 B1, that the composition of end products produced by each individual probiotic strain were specific and different than the other strains present in the mixed culture. For example, each individual strain of the probiotic belonging to multiple mixed strain probiotic culture produced different bacteriocins, therapeutic peptides, short chain fatty acids, and varying quantities of organic acids such as lactic, acetic, butyric and propionic acids. Consequently, the patented invention involved the use of multiple mixed strain probiotics to arrive at complex and mixed immunomodulins to successfully inhibit and prevent or cure Covid-19 infection. Yet no mention was made about probiotic produced memiRNAs and their involvement in the invention.
The patent also mentioned and outlined various routes of administration to stimulate the host immune system through activation of the specific tissues of the organs, such as buccal cavity, throat, lungs, and the gastrointestinal tract etc. In essence, after going through series of the experiments conducted in this investigation, we can better understand and can offer a logical explanation regarding how and why the patented novel invention worked well to inhibit the SARS-CoV-2 virus and other associated secondary pathogenic micro-organisms, proven through conducting RT-PCR test as follows: prokaryotic probiotic bacteria, along with their immunomodulins, might have stimulated human epithelial cells to produce specific microRNAs which can exit out of the eukaryotic epithelial cells into the trachea, bronchi, bronchioles, alveoli and /or buccal cavity and/or the Gastrointestinal tract, depending on the route of administration. These microRNAs produced by human eucaryotic cells (under the influence of the procaryotic probiotic bacteria or perhaps due to the immunomodulins produced by such probiotic bacteria or due to combined effect of probiotics and their immunomodulins) must have exited from the eucaryotic cells either passively due to their smaller size (20-25 nucleotides length) or thru a permease-assisted membrane transport system, to inhibit the pathogenic bacteria and viruses by blocking either their gene transduction or by blocking translation of specific mRNAs coding for an essential protein of such pathogenic bacteria or viruses.
This can be the reason why Microbiota and Microbiome were maintained well with the administration of multiple mixed strain probiotics along with their immunomodulins. Such well-maintained and orchestrated Microbiota might have further stimulated the immune system, specifically to produce T- Regulatory cells through immunomodulation, to control the deadly cytokine storm during COVID-19 infection. In this scenario perhaps it is the combination of probiotics and their immunomodulins (including probiotic produced procaryotic microRNAs or memiRNAs) along with the eucaryotic produced microRNAs (produced under the influence of probiotics), must have altered the Microbiota to assist the immunomodulation, to override the viral, bacterial, and cytokine storm during COVID-19 infection. Although the prokaryotic and eukaryotic microRNAs involvement was not discussed in US patent # 11,077,052 B1, this investigation brought into light their significance and function to arrest the viral and bacterial infections.
In addition to this, microRNAs or vesicular microRNAs produced and released as part of the immunomodulins by the multiple mixed strain probiotics might have entered into the systemic circulation and ultimately ended up in the eucaryotic cell cytoplasm, due to their smaller size (100 to 500 nucleotides length). While they are in the cytoplasm, they might have acted directly to suppress the viral mRNA from translating to produce viral proteins or indirectly induced the eukaryotic cell to produce microRNAs to inhibit the coronavirus multiplication. Now that we have uncovered the presence of probiotic produced RNA fractions (memiRNAs), as part of their immunomodulins, we can conclude or hypothesize stating that the possible reason for the success of the novel patented invention (US Patent # 11,077,052 B1) to prevent or cure the COVID-19 infection can be attributed to microRNAs along with other factors produced by the multiple mixed strain probiotics.
Additionally, the success of our earlier invention of using the multiple mixed strain probiotic therapy to prevent or cure nosocomial infections due to multiple antibiotic resistant micro-organisms (C.diff and MRSA etc.) can also be attributed partly to both the probiotic produced microRNAs (memiRNAs) as well as probiotic induced microRNAs (mimiRNAs) produced by the eukaryotic cells, which subsequently leaked or excreted into the GI tract to inhibit C.diff and MRSA bacteria. Such inhibition was perhaps due to blockage of the translation of the essential growth proteins of pathogenic bacteria by microRNA. In addition to the memiRNAs, probiotics might have competed with the antibiotic-resistant microorganisms through nutritional competency, bacteriocins production and immune stimulation to suppress their growth. In addition, as outlined earlier, the vesicular microRNAs produced by probiotics must have entered into the intestinal epithelial cells thru permease mediated membrane transport system to stimulate the human GI tract epithelial cells to produce eukaryotic microRNAs. Such microRNAs can be emptied into the GI tract as fecal micro RNAs to inhibit nosocomial infection-causing pathogenic bacteria, such as C.diff, MRSA, Enteropathogenic E.coli, members of the Klebsiella and carbapenem resistant Enterococci.
We can also hypothesize that the partial success of fecal microbiota therapy to treat the nosocomial infections was due to the presence of not only probiotic-rich Microbiota and the probiotic produced immunomodulins with the inclusion of probiotic produced microRNAs i.e. memiRNAs, but also due to the presence of probiotic induced eucaryotic cell produced fecal microRNAs. Although fecal microbiota transplant therapy is not safe in the long run, it acts as a quick temporary fix. The superior and safer option is to use defined multiple mixed strain probiotics along with their immunomodulins, or perhaps a combination of fecal microbiota therapy and multiple mixed strain probiotic therapies, with a clear conscious of using predominant fraction of defined probiotics.
Our observation and explanation through experimental proof of the involvement of probiotic produced microRNA (memiRNAs) in the immunomodulins and their effect on stimulating the production of eukaryotic microRNAs (miRNAs), which will eventually exit out of the cells into the lumen to maintain the homeostasis of the intestinal Microbiota and their effect on the host immune system, explains why such a therapeutic approach when used as an adjuvant therapy along with the standard cancer therapies and the immune check point therapy worked very well to cure cancer efficiently with great success, with the least relapse after remission.
Conclusion
The following conclusions were drawn from this investigation:
- The significant feature of this investigation is that the stationary phase of probiotic bacteria (with or without the presence of extra chromosomal genes or plasmids) is controlled by internally produced mega-microRNAs (memiRNAs) to retard excess multiplication of the probiotic bacteria in addition to the other known physiological factors such as nutrient depletion, end product inhibition etc.
- Additionally, the memiRNA produced by probiotic bacteria exits out of the cell and becomes part of the growth end product/ immunomodulin which can inhibit E.coli, but not the parental probiotic bacteria which produced it.
- Furthermore, the mega-microRNA (memiRNA) present in the phage lysate of Streptococcus thermophilus inhibits the growth of various phage-unrelated S. thermophilus probiotic strains. Whereas the immunomodulins produced by the probiotic bacteria without any phage infection did not exhibit such inhibition. It indicates that the mega-microRNAs (memiRNAs) produced during phage infection have different physiological properties.
- It was proven that the S.thermophilus phage lysate had inhibitory RNA particles, evidenced by the fact that such RNAs did not inhibit E.coli after it was inactivated with the use of RNase enzyme. However, such inhibition still existed even after the proteins, peptides, bacteriocins and DNA particles were inactivated using protease, amino peptidase and DNase enzymes, thereby proving that the inhibitory factor was indeed RNA-related mega-microRNA (memiRNA).
- The mega-microRNAs (memiRNAs) produced under the influence of S. thermophilus bacteriophage had significant stimulatory effect on Lactobacillus helveticus probiotic, when S. thermophilus and L. helveticus were grown together. It proves that mega-microRNAs (memiRNAs) produced by probiotic bacteria will exhibit both the prevention of translation of some proteins in certain species, and stimulation of translation of growth proteins in other species.
- Overall, this investigation proved that probiotic bacteria produce microRNAs, which we have labeled as mega-microRNAs or memiRNAs. Their function varies according to their biologically-challenged circumstances encountered in the GI tract, such as undue strain dominance of the microorganisms in microbiota and bacteriophage infections etc.
- After conducting this elaborate research involving several experiments, the following inference was drawn on the validity or the reasoning and scientific rationale behind the success of US patent # 11,077,052 B1, regarding the usage of multiple mixed strain probiotics along with their immunomodulins to prevent or treat SARS-CoV-2 coronaviral Covid-19 infections. The multiple mixed strain probiotic produced immunomodulins must have microRNAs (memiRNAs) as one of the integral ingredients.
- When these preparations were introduced either through nasal passages or into the Gastrointestinal tract, the probiotics and their immunomodulins with memiRNAs must have stimulated the human epithelial cells to produce eukaryotic miRNAs, which will eventually exit into lungs or GI tract lumen as fecal miRNAs. The memiRNAs and miRNAs, along with the probiotics and their immunomodulins and the orchestrated immune system collectively must have inhibited the SARS-CoV-2 corona virus
Disclosure
The author is a scientist with degrees in Veterinary medicine, MS degree in Microbiology, and Ph.D. in Bacteriology, virology and Food Technology from Iowa State University, USA. He is heavily involved in the probiotic research over 50 years, and holds over 150 national and international Patents, and published over 170 research articles. He serves as the president of IMAC Inc. which is involved in the manufacture of food-grade beneficial bacterial cultures and other high-tech enzyme fortified functional products that go into manufacture of various dairy products in the United States, Canada, Europe, Asia, Korea, and South America.
Acknowledgements
I am extremely thankful and grateful to Mr. Venkat (V. R.) Mantha, director of Quality Control and Research and Development of IMAC [ International Media And Cultures ], a division of American Dairy and Food Consulting Laboratories Inc., of Denver, Colorado USA, for coding, critiquing, and compiling the data, and for helping me to prepare this elaborate research manuscript for publication. My sincere appreciation and deep gratitude to Mr. Sridhar Reddy and Rasheed Hussain for their contribution to prepare the tables and graphs that are presented in this research article. Thanks, are also extended to all the individuals and fellow scientists who have participated in this extensive research project.
Conflict of Interest
No conflict of interest.
References
1. Reddy MS. Selected Multiphase Treatment for Coronavirus Respiratory Infections. US Patent # 11, 077, 052 B1. 2021: 1-31.
2. Smolenski DJ. Micro-RNA – A Nobel winning discovery. Natural Sciences 2024-10-18. https: //.portal.umk.pl/en/article/microrna-a-nobel-winning- discovery#:~:text=In%202024%2C%20the%20Nobel%20Prize,post%2Dtranscriptional%20regulation%20of%20genes
3. Wightman I Ha, Ruvkun. Post transcriptional regulation of the heterochronic gene Lin-14 by Lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993; 75: 855-862.
4. Ruvkun G. “ Glimpses of tiny RNA world. (Perspectives: Molecular Biology) “. Science.2001; 294 (5543), No. 5543: 797.
5. Ambros V, Ruvkun G. Recent molecular genetic explorations of Caenorhabditis elegans MicroRNAs. Genetics. 2018; 209 (3): 651-673. – https://doi.org/10.1534/generics.118.300291.
6. Barrel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009; 136: 215-233.
7. Jason MD, Lund AH. MicroRNA and cancer. Med Oncol. 2012; 6 (6): 439-446.
8. Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev enet. 009; 10(10):704.
9. Chen JQ, Papp G, Szodoray P, Zehr M. The role of microRNAs in the pathogenesis of autoimmune diseases. Autoimmun Rev. 2016; 15 (12): 1171-1180.
10. Jinnin M. microRNA in autoimmune disorders. Japanese Journal Of Clinical Immunology. 2011; 34 (6): 439-446.
11. Maes OC, Chertkow HM, Wang E, Schiller HM. MicroRNA: Implications for Alzheimer’s disease and other human CNN disorders. Curr Genomics. 2009; 10(3): 154-168.
12. Chen XM, Huang QC, Yang SL, Chu YL, Yan YH, Han L, et al. Role of Micro RNAs in the pathogenesis of Rheumatoid Arthritis: Novel perspectives based on literature. Medicine. 2015; 94 (31):e 1326.
13. Reddy MS. Mechanism of thrombosis during COVID-19 infection due to SARS-CoV-2 virus and its variants, and a clinically proven strategy to combat with probiotics and their immunomodulins. Medical Research Archives. 2022: 1 – 21. https://esmed.org/MRA/Index.php/mra/article/view/3075
14. Reddy MS. Scientific and medical research on Dr. M.S. Reddy’s Multiple Mixed Strain Probiotic Therapy and its influence on assisting to cure or prevent the nosocomial infections, synergistically enhancing the conventional cancer therapies, as an adjuvant, and its possible potential to prevent or cure COVID -19 novel Coronavirus infection by balancing the intestinal Microbiota and Microbiome through modulation of the human immune system. Int J Pharma Sci Nanotech. 2020; 13(3): 4876- 4906. https: //doi.org/10.37285/ijpsn.2020.13.3.3.
15. Reddy MS. Prevention of Viral Transmission by Naked Genetic Material. US Patent # 11,643,641 B2. 2023: 1-20.
16. Reddy MS, Reddy DRK, Prasad NAV. Herbal and pharmaceutical drugs enhanced with probiotics. US Patent # 6, 080, 401. 2000:1-48.
17. Reddy MS, Vedamuthu ER, Washam CJ, ReinboldGW. Differential agar medium for separating Streptococcus lactis and Streptococcus cremoris. Appl Microbiol. 1969; 198: 755-759.
18. Reddy, MS, Vedamuthu ER, Washam CJ, Reinbold GW. Agar medium for differential enumeration of lactic Streptococci. Appl Microbiol. 1972; 24: 947- 952.
19. Reddy MS, Vedamuthu ER, Reinbold GW. A differential broth for separating the lactic streptococci. J Milk Food Technol. 1971; 34: 43-45.
20. Reddy MS, Vedamuthu ER, Washam CJ, Reinbold GW. Associative growth studies in three-strain mixtures of lactic Streptococci. Appl Microbiol. 1972; 25: 953 – 957.
21. Reddy MS, Reddy DRK. Development of multiple mixed strain probiotics for “Probiotic therapy “under clinical conditions, to prevent or cure the deadly hospital acquired infections due to Clostridium difficile (C. diff) and Methicillin Resistant Staphylococcus aureus (MRSA) Int J Pharma Sci Nanotech. 2016; 9 (3): 3256- 3281.
22. Reddy MS. Probiotics: Genesis, current definition, and proven therapeutic properties. JAAPI. 2021; 1 (2): 18 – 26.
23. Reddy MS. Proven novel method to curtail COVID-19 pandemic, LONG COVID, any further viral pandemics, including influenza due to RNA/DNA viruses, through inactivation of their naked RNA/DNA, and Hospital Acquired [Nosocomial] infections. Arch Neurol & Neuroscience. 2023; 16 (1): 1-20. DOI: 10.33552/ANN 2023. 16. 000880.
24. Reddy MS. Dr. M.S. Reddy’s Multiple Mixed Strain Probiotis Adjuvant Cancer Therapy, to complement immune check point therapy and other traditional cancer therapies, with least autoimmune side effects through eco- balance of human Microbiome. Int J Pharma Sci Nanotech. 2018; 11 (6): 4295- 4317.
25. Reddy MS. Immunomodulatory effect of “ Dr. M.S. Reddy’s Multiple Mixed Strain Probiotic Therapy “, to cure or prevent hospital acquired (Nosocomial) infections due to Clostridium difficile (C. diff), other pathogenic bacteria and autoimmune diseases. Int J Pharma Sci Nanotech 2018; 11(1): 3937- 3949.
26. Reddy MS, Reddy DRK. Therapeutic effect of multiple mixed strain probiotics to prevent or cure the hospital associated infections due to Clostridium difficile – C. diff and methicillin resistant Staphylococcus aureus – MRSA. Presented at the 26 th annual AAPI medical convention (American Association of Physicians of Indian origin). June 26-29 (Las Vegas, Nevada, USA), 2008.
27. Reddy MS, Reddy DRK. Isolation and the determination of the major principle or causative agent behind the 2016 published breakthrough discovery of Dr. M. S. Reddy’s “Multiple Mixed Strain Probiotic Therapy “, in successfully treating the lethal hospital acquired infections due to Clostridium difficile (C. diff) and Methicillin Resistant Staphylococcus aureus (MRSA). Int J Pharma Sci Nanotech. 2016; 9 (6): 3556-3566. https://doi.org/10.37285/ijpsn.2016.9.6.7
28. Reddy MS, Reddy DRK. Therapeutic probiotics for cancer reduction. AAPI journal/ May/ June. 2015: 55-56.
29. Reddy MS, Reddy DRK. Probiotics, biotechnology, and Ayurvedic herbs – complementary and alternative medicine. AAPI Journal / May/ June. 2004: 32-33.
30. Reddy MS, Reddy, DRK. Anti-aging: Review and experimental clinical study of bioavailable calcium – probiotics and their effect on reversing osteopenia, osteoporosis, and other common health conditions. Int J Pharma Sci Nanotech. 2011; 4: 1436 – 1444.
31. Reddy MS. Mechanism of Cytokine Storm in COVID-19: How can probiotics combat it? JAAPI (Journal of the American Physicians of Indian Origin). 2021; 1 (3): 27-35.
32. Voinnet O. Original, biogenesis, and activity of plant microRNAs. Cell. 2009; 136: 669-687.
33. Nogales AR, Algieri F, Garrido-Mesa J, Vezza T, Utrilla MP, Chueca N, et al. The administration of Escherichia coli Nissle 1917 ameliorates development of DSS – induced colitis in mice. Front Pharmacol. 2018; 9: 468.
34. Davoodvandi A, Marzban H, Goleij P, Sahebkar A, Morshedi K, Rezaei S, et al. Effects of therapeutic probiotics on modulation of microRNAs. Cell Communication And Signaling. 2021; 1934: 1-22. https:/doi.org/10.1186/s12964-020-00668-w .
35. Brogna C, Cristoni S, Petrillo M, Bisaccia Dr, Lauritano F, Montano L, et.al. The first report on detecting SARS-CoV-2 inside bacteria of the human gut microbiome: A case series on Asymptomatic family members and a child with Covid-19. F1000 Res. 2024 Oct 16; 11:135. Doi: 10.12688/f1000 research. 77421.3. PMCID: PMC 11502994 PMID: 39464247.

