Detection of SARS-CoV-2 Omicron BA.4 and BA.5 in Cameroon
Initial detection of SARS-CoV-2 Omicron BA.4 and BA.5 sub variants associated with the onset of the fifth wave of COVID-19 in Cameroon between December 2021 and June 2022: phylogenetic and whole genome analysis
John Otokoye Otshudiema1,2, René Ghislain Essomba3,4, Moussa Moise Diagne5, Valentina Josiane Ngo Bitoungui3,6, Yvette Ebogo3, Eric Minkala3, Emmanuel Lofiko Lokilo7, Karl Njuwa Fai8, Sara Eyangoh9, Selassie Kumordjie10, Dorine Ngono1, Moise Henri Moumbeket9, Placide Mbala Kingebeni2,7, Steve Ahuka Mundeke2,7, Nicksy Gumede11, Joseph Fokam12,13, Linda Esso4,12, Louis Richard Njock4,12, Georges Alain Etoundi Mballa12, Richard Njouom9, Yap Boum II4,12 Marie Claire Assoumou Okomo3,4
- COVID-19 Genomic Surveillance, Incident Management System Team, World Health Organization (WHO), Yaounde, P.O. Box 155 Yaounde, Cameroon
- Kinshasa School of Public Health and Faculty of Medicine, University of Kinshasa, Kinshasa I, P.O. Box 190 Kinshasa XI, Democratic Republic of the Congo
- Genomic Surveillance Department, National Public Health Laboratory (NPHL) of Cameroon, Ministry of Public Health, Yaounde I, Cameroon
- Faculty of Medicine and Biomedical Sciences, University of Yaounde 1, P.O. Box 337 Yaounde, Cameroon
- WHO Reference Sequencing Laboratory, Virology and Genomic Surveillance, Institut Pasteur de Dakar, 12900 Dakar, Senegal
- Faculty of Medicine and Pharmaceutical Sciences, University of Dschang, P.O. Box 96 Dschang, Cameroon
- WHO Reference Sequencing Laboratory, Epidemiology and Global Health, Institut National de Recherche Biomédicale (INRB), Kinshasa, Kin I Gombe, Democratic Republic of Congo (DRC)
- Epicentre Médecins Sans Frontières (MSF), Yaounde I, Cameroon
- Virology Unit, Centre Pasteur du Cameroun (CPC), Yaounde, B.P. 1274 Yaounde, Cameroon
- Department of Parasitology, Noguchi Memorial Institute for Medical Research, Accra, GA-337-3045, Ghana
- COVID-19 Incident Management Support Team, WHO Regional Office for Africa, World Health Organization, Brazzaville, P.O. Box 06 Brazzaville, Republic of Congo
- Virology Service, Centre International de Recherche Chantal Biya (CIRCB), Yaounde, P.O. Box. 3077 Yaounde, Cameroon
- National COVID-19 Incident Management Team, Public Health Emergency Operations and Coordination Center, Ministry of Health, Yaounde I, Cameroon
- Faculty of Health Sciences, University of Buea, P.O. Box 63 Buea, Cameroon
†These authors contributed equally as first authors.
††These authors jointly supervised this work as senior authors.
OPEN ACCESS
PUBLISHED: 31 January 2025
CITATION: Otshudiema, JO., Essomba, RG., et al., 2025. Initial detection of SARS-CoV-2 Omicron BA.4 and BA.5 sub variants associated with the onset of the fifth wave of COVID-19 in Cameroon between December 2021 and June 2022: phylogenetic and whole genome analysis. Medical Research Archives, [online] 13(1). https://doi.org/10.18103/mra.v13i1.6155
COPYRIGHT: © 2025 European Society of Medicine. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
DOI: https://doi.org/10.18103/mra.v13i1.6155
ISSN: 2375-1924
ABSTRACT
Background: Two sub variants (BA.4 and BA.5) of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant are concerning as they are spreading rapidly worldwide; however, no published data concerning these variants are available in Cameroon. We report the early detection of these new sub variants that are associated with the onset of the fifth wave of coronavirus 2019 (COVID-19) in Cameroon.
Methods: Positive samples were selected for next-generation sequencing (NGS). BA.4 and BA.5 complete genome sequences underwent sequence data analysis, epidemiology analysis of COVID-19’s resurgence and wave, recombination and pairwise matrix analysis, and phylogenetic analysis. We selected the first nine SARS-CoV-2 Omicron BA.4 and BA.5 sub variants detected in Cameroon using local whole genome sequencing for the NGS analysis.
Results: During the fifth wave of resurgence of COVID-19 cases in Cameroon, it was found that the Northwest and Littoral regions were the most affected areas, while the Center and Littoral regions recorded the highest number of new deaths. The study identified evidence of recombination between the BA.2 sub variant and BA.4 and BA.5 Cameroonian strains. This result highlights the dynamic nature of SARS-CoV-2 evolution. The BA.5 strain (entitled hCoV-19/Cameroon/23850/2022) showed the highest sequence similarity to the first reported genome of the Omicron strain with 497 mutations. Phylogenetic analysis revealed that these nine Omicron sub variants were grouped into a distinct and highly distant cluster separate from the first Omicron variant detected in Botswana and were intermixed with sequences from other countries (the United States, Denmark, Scotland, and England), thus implying multiple introductions of the BA.4 and BA.5 sub variants in Cameroon.
Conclusions: Omicron BA.4 and BA.5 sub-lineages are associated with the onset of the fifth wave of COVID-19 in Cameroon. In addition to providing early warning of COVID-19 resurgence, continuous local genome sequencing of emerging variants is essential for detecting variants of concern, thereby guiding the country’s response. This study emphasizes the value of real-time surveillance.
Keywords
COVID-19; Omicron BA.4 and BA.5 sub-variants; fifth wave; whole genome analysis.
THE EUROPEAN SOCIETY OF MEDICINE
Medical Research Archives, Volume 13 Issue 1
RESEARCH ARTICLE
Introduction
Emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants has fundamentally shaped coronavirus 19 (COVID-19) epidemiology globally and is of particular significance in Africa. Cameroon reported its first COVID-19 cases on March 5, 2020 followed by four distinct pandemic waves that were associated with successive variants of concern (VOC). By mid-2022, the Omicron variant and its sub-lineages achieved global dominance, representing 84% of sequences submitted to the Global Initiative for Sharing All Influenza Data (GISAID).
The evolution of Omicron has been characterized by successive sub-lineages beginning with BA.1–3 during South Africa’s fourth wave (November 2021–January 2022). Subsequently, BA.4 and BA.5 sub-lineages emerged and drove South Africa’s fifth wave (April–June 2022) as these sub-lineages demonstrated enhanced transmissibility. These variants significantly impacted healthcare systems, particularly by causing an increase in hospitalizations among elderly populations.
Genetic analysis revealed that BA.4 and BA.5 possess distinct molecular signatures as these two sub-lineages have 33 and 34 spike mutations, respectively. These sub variants present a closer genetic relationship to BA.2 than BA.1 and harbor unique spike protein mutations (L452R and F486V) that enhance their immune evasion capabilities. Recent studies have demonstrated that BA.5 shows an increased resistance to neutralizing antibodies compared to its predecessors.
The World Health Organization (WHO) defines COVID-19 resurgence as a spike in new cases following minimal transmission for at least two weeks and establishes a framework with three critical phases: (1) resurgence response, (2) control, and (3) alert. This framework guides regional preparedness and intervention strategies on a global level.
While recent surveillance in Cameroon (December 2022–March 2023) has indicated the emergence of newer variants, including XBB.1, BQ.1.1, and atypical BA.4.6/XBB.1 recombinants, our crucial study focuses on the initial emergence and characterization of BA.4 and 5. This study provides essential baseline data for understanding the evolutionary trajectory of SARS-CoV-2 in Cameroon and contributes to a broader understanding of variant emergence patterns in Africa. Although WHO declared an end to COVID-19 as a global health emergency in May 2023, understanding the evolutionary dynamics of SARS-CoV-2 variants remains critically important for several reasons: (1) it helps predict future viral behavior patterns, (2) informs preparedness strategies for potential resurgences and (3) provides valuable insights for managing emerging respiratory pathogens. Furthermore, this historical analysis of variant emergence and spread patterns serves as a crucial reference point for developing improved surveillance systems and response mechanisms for addressing future pandemic threats.
This study aimed to analyze the phylogenetic relationships between Cameroon’s BA.4/5 sub-lineages and the original Omicron variant, evaluate their correlation with Cameroon’s fifth COVID-19 wave, and compare whole genome sequences with international data to understand viral transmission dynamics.
Methods
RATIONALE FOR METHODOLOGY
The methodological framework for this study was carefully selected to ensure comprehensive characterization of SARS-CoV-2 variants while maintaining analytical rigor. We selected an integrated approach that combined molecular diagnostics, next-generation sequencing (NGS), and phylogenetic analysis for several key reasons: (1) real-time polymerase chain reaction (PCR) provided rapid initial screening with high sensitivity, (2) NGS enables detailed genomic characterization with superior depth coverage, and (3) phylogenetic analyses allowed robust evolutionary relationship determination. This multi-faceted approach aligns with WHO recommended protocols for SARS-CoV-2 surveillance while concurrently enabling detailed variant tracking and characterization.
STUDY DESIGN AND POPULATION
This retrospective observational study analyzed SARS-CoV-2-positive cases that had been recorded between December 2021 and June 2022. We chose this design to enable comprehensive genomic surveillance while minimizing selection bias. Data were obtained from the National Public Health Laboratory and Centre Pasteur du Cameroon (NHL and CPC) databases, respectively, along with the national line listing.
SAMPLE COLLECTION AND PROCESSING
Following standardized national COVID-19 protocols, nasopharyngeal and oropharyngeal specimens were collected from three strategically selected cohorts: (1) symptomatic individuals with acute respiratory infection manifestations, (2) high-risk contacts of confirmed cases, and (3) international travelers. This sampling strategy was designed to capture both community transmission and potential introduction of new variants.
SARS-CoV-2 MOLECULAR DIAGNOSTICS
We selected the DAAN Gene extraction kit for RNA extraction due to its proven reliability and standardization capabilities. Real-time amplification targeted the viral ORF1ab and N gene using TaqMan probes, which had been chosen for their high specificity and sensitivity. The established threshold values (Ct < 30 for positive, Ct > 30 for negative) were based on validated protocols that ensured optimal diagnostic accuracy.
NEXT-GENERATION SEQUENCING
The choice of the Illumina COVIDSeq protocol was based on its Food and Drug Administrations (FDA) approval status and demonstrated high-throughput capabilities. This method generates 400bp amplicons through multiplexed PCR reactions and provides optimal coverage for variant detection. Library preparation procedures were standardized using IDT for Illumina PCR Unique Dual Indexes to minimize index hopping and ensure accurate demultiplexing.
Sequence Analysis and Quality Control
The EDGE COVID-19 bioinformatics platform was selected for its comprehensive analysis capabilities and validated workflow. Quality control measures included stringent parameters for trimming, alignment, and variant calling so as to ensure reliable genomic data. Lineage assignment utilized the latest versions of Pangolin COVID-19 software and NextClade for accurate variant classification.
PHYLOGENETIC AND GENOMIC ANALYSIS
Maximum likelihood methods based on the Hasegawa–Kishino–Yano model were chosen for their robust statistical framework and capability for handling large datasets. Tree visualization and recombination detection methods were selected based on their proven reliability in SARS-CoV-2 genomic analyses.
Results
CHARACTERISTICS OF BA.4 AND BA.5 SUB-VARIANT STRAINS
In this study, we detected 381 COVID-19 positive cases that had been tested by reverse transcriptase polymerase chain reaction (RT-PCR). Of 381 positive cases, 318 (83.5%) full genomic sequences were obtained. The genome sequences range from 25 to 28.9 kb. Among the 318 complete sequences, the Omicron variant was predominant with 299 (94.0%) sequences: (1) 236 (78.9%) BA.1; (2) 26 (8.7%) BA.2; (3) 0 (0.0%) BA.3; (4) 3 (1.0%) BA.4; and (5) 6 (2.0%) BA.5. Other Omicron sub-lineages were also found: (1) 28 (9.4%) XG, (2) XP, and (3) other recombinants. Interestingly, this study presents the first report of BA.4 and BA.5 sub variants in Cameroon. Regarding BA.4 and 5 sub-variant strains, the purpose of sampling was travel (six strains) and routine screening (three strains). Eight strains were isolated from Cameroonian patients and one strain from a Congolese patient, all of which were asymptomatic. Five strains were isolated from unvaccinated patients, whereas two strains were detected among vaccinated patients. The vaccination status of the two other cases was unknown.
| N | Sample ID | Location | Sample Collection Date | Specimen Type | Purpose of Sampling | If travel, provenance/destination | Nationality/residence | Age (y) | Gender | Symptoms status | Vaccination status/vaccine-dose | qPCR Result Date | SARS-CoV-2 PCR Result | CT Value Gene N | CT Value ORF-1ab | Clade | Nextclade_pango |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HCY–003411 | Africa/Cameroon/Littoral/Deido | 06/06/2022 | Nasopharyngeal and oropharyngeal swabs | Travel | Entry from Addis Ababa – Ethiopia | Cameroonian/Cameroon | 38 | Male | Asymptomatic | Unvaccinated | 06/06/2022 | Positive | 23.9 | 25.6 | 22B (Omicron) | BA.5 |
| 2 | B-126495 | Africa/Cameroon/Littoral/Deido | 07/06/2022 | Nasopharyngeal and oropharyngeal swabs | Travel | Entry from Milan – Italy | Camerounian/Cameroon | 40 | Male | Asymptomatic | Unvaccinated | 07/06/2022 | Positive | 21.11 | 23.5 | 22B (Omicron) | BA.5 |
| 3 | HCY–16401 | Africa/Cameroon/Littoral/Bangue | 02/01/2022 | Nasopharyngeal and oropharyngeal swabs | Travel | Entry from Lomé – Togo | Cameroonian/Cameroon | 56 | Male | Asymptomatic | Unvaccinated | 02/01/2022 | Positive | 22.5 | 25.6 | 22B (Omicron) | BA.5 |
| 4 | HCY-49830 | Africa/Cameroon/West/Mifi | 09/01/2022 | Nasopharyngeal and oropharyngeal swabs | Travel | Exit from Mifi – West Cameroon | Cameroonian/Cameroon | 38 | Female | Asymptomatic | Unvaccinated | 09/01/2022 | Positive | 30.8 | 25.9 | 22B (Omicron) | BA.5 |
| 5 | 332411 | Africa/Cameroon/South/Kribi | 24/12/2021 | Nasopharyngeal and oropharyngeal swabs | Travel | Exit from Kribi – South Cameroon | Italian/Cameroon | 62 | Male | Asymptomatic | Vaccinated – AstraZeneca – 2 doses | 24/12/2021 | Positive | 20.3 | 18.5 | 22B (Omicron) | BA.5 |
| 6 | SUD-23850 | Africa/Cameroon/South/Kribi | 12/06/2022 | Nasopharyngeal and oropharyngeal swabs | Routine screening | Not applicable | Cameroonian/Cameroon | 36 | Female | Unknown | Unknown | 12/06/2022 | Positive | 17.5 | 22.3 | 22B (Omicron) | BA.5 |
| 7 | 0328873 | Africa/Cameroon/Center/Cité verte | 20/04/2022 | Nasopharyngeal and oropharyngeal swabs | Routine screening | Not applicable | Cameroonian/Cameroon | 31 | Male | Unknown | Unknown | 20/04/2022 | Positive | 32 | 35.2 | 22A (Omicron) | BA.4.1 |
| 8 | B-349629 | Africa/Cameroon/Center/Cité verte | 13/06/2022 | Nasopharyngeal and oropharyngeal swabs | Routine screening | Not applicable | Cameroonian/Cameroon | 35 | Female | Asymptomatic | Unvaccinated | 13/06/2022 | Positive | 15.5 | 19.4 | 22A (Omicron) | BA.4.1 |
| 9 | B-082189 | Africa/Cameroon/Center/Djoungolo | 29/05/2022 | Nasopharyngeal and oropharyngeal swabs | Travel | Exit from Yaoundé Cameroon to Kinshasa, DRC | Congolese (DRC)/DRC | 42 | Male | Asymptomatic | Vaccinated – J&J – 1 dose | 29/05/2022 | Positive | 20.3 | 17.5 | 22A (Omicron) | BA.4.1 |
EPIDEMIOLOGICAL ANALYSIS AND LINK WITH RESURGENCE OF CASES
In terms of new cases, the study revealed that the Northwest (890 new cases) and Littoral (678 new cases) regions were the most affected areas during the last six weeks of the study period (from Epidemiological Week (EW) 28 through EW 33 in 2022). While the Central and Littoral regions presented the highest number of new deaths (four cases), the highest percentage of change in new cases over the last six weeks following the end of the study period was detected in the Northwest and East regions (1870% and 700%, respectively).
| Region | Cumulative cases | New cases in the last 6 weeks (EW28-EW33) | New cases in the last 3 weeks (EW31-33) | New cases in the 3 previous weeks (EW31-33) | % Change in new cases in last 6 weeks | Cumulative deaths | Total of new deaths in the last 6 weeks (EW28-EW33) | New deaths in the last 3 weeks (EW31-33) | New deaths in the 3 previous weeks (EW31-33) | % Change in deaths cases in last 6 weeks |
|---|---|---|---|---|---|---|---|---|---|---|
| Adamawa | 4,153 | 47 | 38 | 9 | 322% | 58 | 0 | 0 | 0 | 0% |
| Centre | 38,193 | 159 | 113 | 46 | 146% | 517 | 4 | 3 | 1 | 200% |
| East | 5,428 | 45 | 40 | 5 | 700% | 84 | 0 | 0 | 0 | 0% |
| Far-North | 2,738 | 2 | 2 | 0 | >100% | 64 | 0 | 0 | 0 | 0% |
| Littoral | 34,565 | 678 | 533 | 145 | 268% | 373 | 4 | 3 | 1 | 200% |
| North | 2,168 | 3 | 3 | 0 | >200% | 43 | 0 | 0 | 0 | 0% |
| North-West | 12,409 | 890 | 847 | 43 | 1870% | 366 | 3 | 3 | 0 | >200% |
| West | 11,825 | 200 | 165 | 35 | 371% | 269 | 0 | 0 | 0 | 0% |
| South | 5,541 | 59 | 42 | 17 | 147% | 73 | 0 | 0 | 0 | 0% |
| South-West | 5,355 | 77 | 68 | 9 | 656% | 88 | 0 | 0 | 0 | 0% |
| Total | 122,375 | 2,160 | 1,851 | 309 | 499% | 1,935 | 11 | 9 | 2 | 350% |
When the peak incidence parameter was considered, none of the three scenarios (A-nationwide, B-Northwest region, and C-South region) exceeded the COVID-19 resurgence–response criterion of 30% greater than the previous peak (fourth wave). During the given period, the average number of cases during the peak nationwide decreased by 69% (723 versus 2,358 cases), whereas increases were observed in the Northwest and Littoral regions of 26% (379 versus 510 cases) and 23% (231 versus 188 cases), respectively. When considering a 7-day moving average, incident cases between EW 29 and EW 31–2022 significantly increased; on average, they rose by 109% nationwide, 375% in the Northwest region, and 60% in the Littoral region. At the national and regional levels, this increase was classified as a “resurgence alert.”
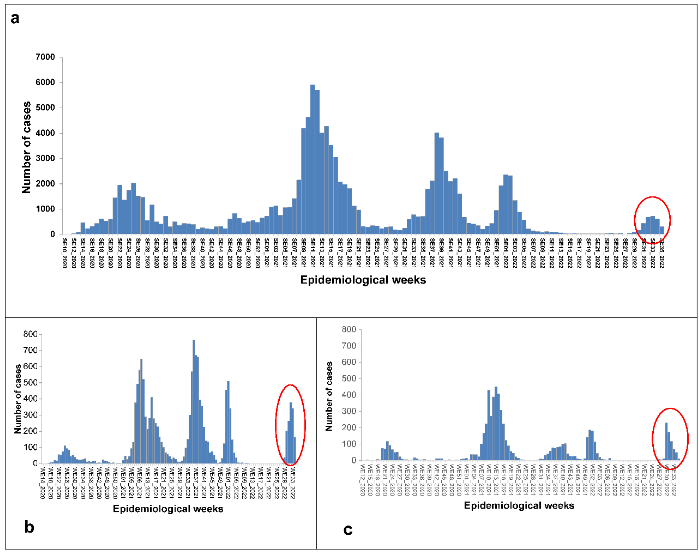
Selected indicators for monitoring resurgence of COVID-19 cases in Cameroon from EW27 to EW33–2022 Following nearly 20 epidemiological weeks of low incidence (control phase), Cameroon has been on resurgence alert since EW31. An increasing trend in the number of new cases from EW28 through EW33 with an increase of 82.7% between EW 28 and 29, 70.5% between EW 29 and EW 30, 173.5% between EW 30 and 31, 54.6% between EW 31 and 32, and 5.5% between EW 32 and 33 has been detected. Most of the sequenced BA.4 and 5 sub-lineages originated from EW 23 and 30. The highest increases in new cases occurred in EW 28 (188.9%) and EW 31 (188.9% and 173.5%, respectively).
| Characteristics | EW27 | EW28 | EW29 | EW30 | EW31 | EW32 | EW33 |
|---|---|---|---|---|---|---|---|
| # new confirmed cases | 18 | 52 | 95 | 162 | 443 | 685 | 723 |
| % change in new cases in last 7 days | -58.1% | 188.9% | 82.7% | 70.5% | 173.5% | 54.6% | 5.5% |
| # new cases per week per million inhabitants | 0.6 | 1.8 | 3.4 | 5.8 | 15.9 | 23.3 | 26.0 |
| # tests performed | 2,691 | 2,226 | 3,556 | 4,282 | 3,379 | 4,774 | 2,816 |
| Test positivity rate | 0.7% | 2.3% | 2.7% | 3.8% | 13.1% | 14.3% | 25.7% |
| # tests performed per 10,000 habitants per week | 0.9 | 0.8 | 1.3 | 1.5 | 1.2 | 1.7 | 1.0 |
| # new deaths | 0 | 0 | 1 | 1 | 0 | 2 | 6 |
| # cases hospitalized | 4 | 7 | 13 | 16 | 35 | 30 | 42 |
| # hospitalized cases under oxygen (ICU) | 0 | 1 | 1 | 1 | 4 | 7 | 13 |
| Detection of Omicron sub variants (BA.4, BA.5) | Yes | Yes | BA.4, BA.5 | Yes | Yes | Yes | Yes |
RECOMBINATION ANALYSIS
Recombination analysis using the Sc2rf pipeline showed evidence of recombination between the BA.2 sub-variant and eight Cameroonian strains.
PAIRWISE COMPARISON MATRIX
The pairwise comparison matrix shows that both BA.4 and BA.5 Cameroonian variants differ significantly from the first reported Omicron variant. The matrix indicates a range of mutations (from 497 to 21,527), thus distinguishing these sub variants from the original strain. The hCoV-19/Cameroon/23850/2022 strain revealed the highest sequence similarity (98.68%) to the reference strain with 497 mutations, while the hCoV-19/Cameroon/332411/2021 strain was the most divergent Omicron strain circulating in Cameroon (44.65%) with 21,527 mutations.
| Gisaid Sample ID | SIN | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hCoV-19/Botswana/R40B59_BHP_3321001248/2021 | 1 | 905 | 3 | 800 | 530 | 9 | 769 | 523 | 497 | 2152 | 7 | 1784 | 2160 | 853 | ||||||
| hCoV-19/Cameroon/003411/2022 | 2 | 76.5 | 3 | 887 | 8 | 826 | 3 | 880 | 2 | 912 | 1 | 918 | 2 | 1417 | 2 | 7379 | 7054 | 8753 | ||
| hCoV-19/Cameroon/0328873/2022 | 3 | 97.9 | 76.7 | 7 | 551 | 9 | 230 | 363 | 347 | 2170 | 5 | 1524 | 2006 | 413 | ||||||
| hCoV-19/Cameroon/126495/2022 | 4 | 85.5 | 7 | 70.8 | 7 | 85.4 | 1 | 545 | 2 | 560 | 6 | 565 | 8 | 1806 | 2 | 4475 | 4071 | 5351 | ||
| hCoV-19/Cameroon/16401/2022 | 5 | 97.9 | 9 | 76.8 | 5 | 98.6 | 8 | 85.4 | 6 | 405 | 383 | 2164 | 3 | 1488 | 1956 | 345 | ||||
| hCoV-19/Cameroon/349629/2022 | 6 | 98.6 | 76.7 | 9 | 98.9 | 2 | 85.6 | 3 | 98.8 | 146 | 2194 | 7 | 1781 | 2091 | 380 | |||||
| hCoV-19/Cameroon/23850/2022 | 7 | 98.6 | 8 | 76.8 | 3 | 99.0 | 2 | 85.6 | 9 | 98.9 | 5 | 99.6 | 2201 | 4 | 1857 | 2168 | 438 | |||
| hCoV-19/Cameroon/332411/2021 | 8 | 44.6 | 5 | 42.9 | 5 | 44.5 | 7 | 42.4 | 8 | 44.6 | 1 | 44.6 | 44.6 | 1 | 2018 | 9 | 1987 | 8 | 2159 | 4 |
| hCoV-19/Cameroon/082189/2022 | 9 | 94.8 | 9 | 76.7 | 5 | 95.2 | 7 | 84.8 | 2 | 95.2 | 2 | 95.2 | 4 | 95.2 | 3 | 44.5 | 8 | 1120 | 1441 | |
| hCoV-19/Cameroon/082216/2022 | 10 | 93.9 | 4 | 76.7 | 7 | 94.2 | 8 | 84.9 | 4 | 94.2 | 3 | 94.4 | 7 | 94.4 | 5 | 44.5 | 8 | 93.5 | 1869 | |
| hCoV-19/Cameroon/49830/2022 | 11 | 97.7 | 7 | 76.8 | 4 | 98.3 | 8 | 85.5 | 1 | 98.4 | 7 | 98.7 | 6 | 98.8 | 1 | 44.6 | 95.2 | 1 | 94.2 | 7 |
PHYLOGENETIC ANALYSIS
The maximum likelihood (ML) phylogeny tree was obtained using the Molecular Evolutionary Genetics Analysis (MEGA) software and showed that the SARS-CoV-2 Cameroonian strains were phylogenetically clustered according to their geographical source. Moreover, Omicron BA.4 and 5 sub variants formed a distinct and highly distant cluster separate from the SARS-CoV-2 reference sequence MN908947.3 (Wuhan-Hu-1).
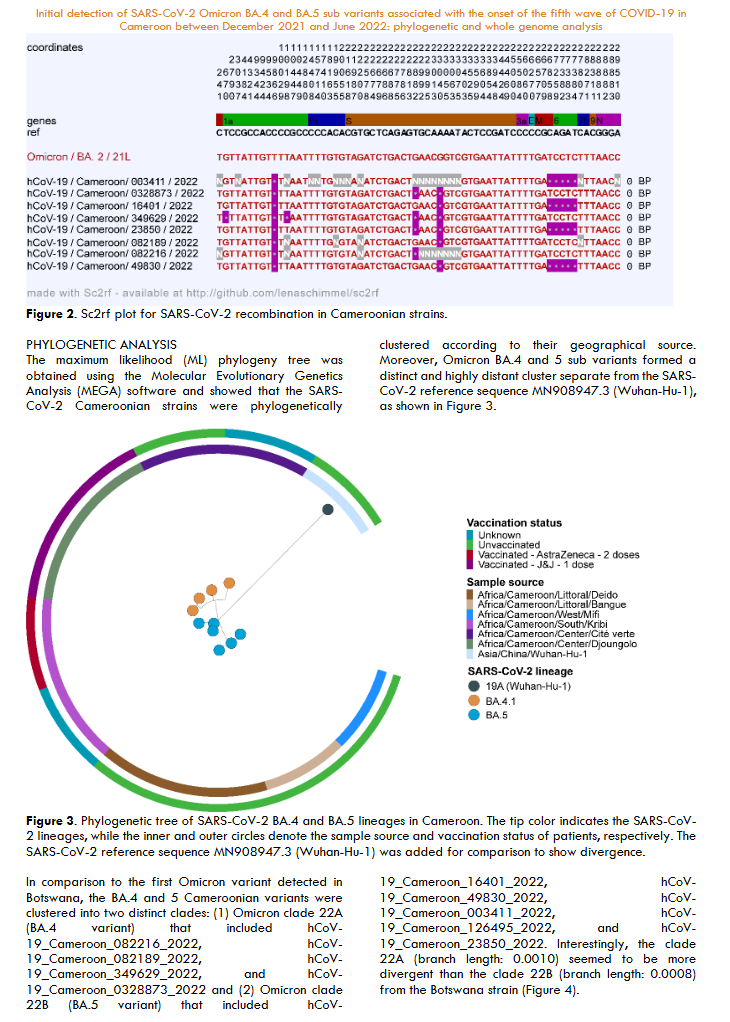
In comparison to the first Omicron variant detected in Botswana, the BA.4 and 5 Cameroonian variants were clustered into two distinct clades: (1) Omicron clade 22A (BA.4 variant) that included hCoV-19_Cameroon_082216_2022, hCoV-19_Cameroon_082189_2022, hCoV-19_Cameroon_349629_2022, and hCoV-19_Cameroon_0328873_2022 and (2) Omicron clade 22B (BA.5 variant) that included hCoV-19_Cameroon_16401_2022, hCoV-19_Cameroon_49830_2022, hCoV-19_Cameroon_003411_2022, hCoV-19_Cameroon_126495_2022, and hCoV-19_Cameroon_23850_2022. Interestingly, the clade 22A (branch length: 0.0010) seemed to be more divergent than the clade 22B (branch length: 0.0008) from the Botswana strain.

Discussion
Our comprehensive genomic and epidemiological analysis of BA.4 and 5 sub-lineages in Cameroon provides crucial insights into SARS-CoV-2 variant evolution and transmission patterns. The findings reveal significant implications for public health strategies while highlighting important considerations for future surveillance efforts.
Emergence and Transmission Dynamics
Identification of BA.4 and 5 sub-lineages in Cameroon, which coincide with the country’s fifth COVID-19 resurgence, reveals a complex transmission landscape. Our analysis identified two primary introduction routes: (1) international travel (44.4%) and (2) local transmission (55.6%), both of which align with patterns observed across Africa. The predominance of BA.5 (32.5%) over BA.2 (20.51%) and BA.4 (11.54%) reflects global trends and suggests enhanced transmissibility of these variants. This pattern of variant displacement provides valuable insights into viral fitness advantages and transmission dynamics.
GENOMIC EVOLUTION AND VARIANT CHARACTERISTICS
A thorough genomic analysis revealed significant divergence among Cameroonian isolates, high contain distinct nucleotide signatures compared to the original Omicron variant. The observed recombination events between BA.2 and BA.4/5 variants, particularly the breakpoint between E and M genes, demonstrate ongoing viral evolution. Phylogenetic analysis showed clear geographic clustering and multiple introduction events and contributes to our understanding of variant dispersal patterns. These findings align with current models of variant emergence and establishment across the African continent.
PUBLIC HEALTH IMPLICATIONS AND RESPONSE STRATEGIES
Our findings present substantial implications for public health strategies. The multiple introduction routes that were identified necessitate enhanced border screening protocols and strengthened local transmission control measures. The immune escape characteristics of BA.4/5 variants in combination with current vaccination coverage suggest potential vulnerabilities in population immunity. These insights are crucial for guiding vaccination strategies and public health planning, particularly in regions with limited healthcare resources.
SURVEILLANCE SYSTEM RECOMMENDATIONS
The study emphasizes the critical need for enhanced monitoring systems in Cameroon and across Africa. Our experience demonstrates that successful variant tracking requires integration of molecular and epidemiological data. While the WHO-AFRO network has been instrumental, our results indicate the necessity for expanded local sequencing capacity. The current recommendation to sequence positive cases requires substantial infrastructure development and technical expertise and supports the establishment of regional sequencing hubs.
STUDY LIMITATIONS AND FUTURE DIRECTIONS
Several important considerations warrant attention when interpreting our findings. The sample size of BA.4 and 5 sequences potentially affects the generalizability of phylogenetic analyses, though statistical validation supports the robustness of our conclusions. The temporal distribution of sampling, while appropriate for initial variant detection, may not fully capture the dynamic nature of variant evolution. Additionally, incomplete clinical outcome data for some cases restricted our assessment of variant pathogenicity.
FUTURE RESEARCH IMPLICATIONS
Moving forward, research efforts should focus on implementing systematic sampling strategies with broader geographic coverage and longer temporal spans for evolutionary analysis. Enhanced clinical data collection protocols will strengthen our understanding of variant dynamics and enable more robust phylogenetic analyses. The establishment of sustainable sequencing capacity in Africa remains crucial and requires continued international collaboration and resource allocation. These improvements will significantly contribute to our understanding of SARS-CoV-2 variant dynamics in Central Africa while supporting the implementation of early warning systems and international coordination in variant monitoring.
Conclusions
Our comprehensive genomic and epidemiological investigation yielded three principal findings with significant implications for SARS-CoV-2 surveillance in Africa. First, we successfully identified and characterized BA.4 and 5 sub-lineages in Cameroon during the country’s fifth COVID-19 wave and then revealed nine distinct sub-lineages with a clear dual transmission pattern (44.4% international travel, 55.6% local transmission). Second, the predominance of BA.5 (32.5%) over other variants, combined with unique genomic signatures and recombination events, provides compelling evidence for ongoing SARS-CoV-2 evolution within the African context. Third, our integrated molecular-epidemiological approach established a robust framework for variant surveillance in resource-limited settings.
These findings have immediate practical implications. The necessity for sustained local genomic sequencing capacity is of utmost importance, followed by the importance of integrated surveillance systems combining molecular and epidemiological data. Additionally, the critical role of international collaboration in variant monitoring and the need for rapid response mechanisms to emerging variants has been clearly demonstrated through our findings.
Looking ahead, our research indicates several crucial developments needed in the field. The implementation of systematic sampling strategies with broader geographic coverage represents an essential next step. This step should be accompanied by the development of sustainable regional sequencing hubs and enhancement of cross-border surveillance networks. Strengthening of data-sharing platforms will further support these initiatives to ensure comprehensive variant monitoring across the continent.
Our results underscore the urgent need for sustained investment in African genomic surveillance infrastructure while providing a practical framework for future variant monitoring efforts. This work contributes to the global understanding of SARS-CoV-2 evolution and establishes a foundation for improved pandemic preparedness in Africa.
Declarations
ETHICS APPROVAL AND CONSENT TO PARTICIPATE:
This operational research was conducted with approval from Cameroon’s Ministry of Public Health as part of the national COVID-19 surveillance program (N°368/NS/MINSANTE/SG/CCOUSP/CSO). This study was also approved by the Cameroon National Ethics Committee for Research in Human Health (N°2020/05/1224/CE/CNERSH/SP). This National Ethics Committee and the Ministry of Public Health waived the need for informed consent. The need for informed consent was waived by the Comité National d’Ethique de la Recherche pour la Santé Humaine [National Ethics Committee for Research in Human Health]. All methods were carried out in accordance with the Declaration of Helsinki.
CONSENT FOR PUBLICATION:
Not applicable.
AVAILABILITY OF DATA AND MATERIALS:
ALL data generated or analyzed during this study are included in this published article.
COMPETING INTERESTS:
The authors declare that they have no competing interests.
FUNDING:
This operational research was funded by the World Health Organization with funds from the United States Government and the African Development Bank through the Central African Economic and Monetary Community as part of the COVID-19 response efforts in Cameroon. The Ministry of Public Health also provided resources. The funders had no role in the design of the study or the collection, analysis, or interpretation of data or in writing the manuscript.
AUTHOR CONTRIBUTIONS:
Conceptualization, J.O.O., R.G.E., M.M.D., E.L.L., K.F., P.M.K, S.A.M., N.G., R.N., L.R.N., Y.B., and M.C.A.O.; Methodology, J.O.O. R.G.E., M.M.D., E.L.L., E.M., S.E., M.H.M., D.G., S.K., P.M.K, S.A.M., N.G., and J.F.; Software, E.L.L., and N.G.; Validation, J.O.O., R.G.E., M.M.D., E.L.L., S.E., S.K., L.E., and N.G.; Formal Analysis, J.O.O., R.G.E., M.M.D., E.L.L., E.M., S.E., N.G., S.K., V.J.B., R.N., L.R.N., Y.E., and M.C.A.O.; Investigation, J.O.O., R.G.E., M.M.D., E.L.L., E.M., S.E., P.M.K, S.A.M., N.G., J.F., S.K., V.J.B., Y.B., L.E., G.A.E.M., M.H.M., R.N., and M.C.A.O.; Resources, J.O.O., and M.M.D.; Data Curation, J.O.O., R.G.E., M.M.D., E.L.L., D.N., P.M.K, S.A.M., and N.G.; Writing – Original Draft Preparation, J.O.O., R.G.E., M.M.D., E.L.L., N.G., and K.F.; Writing – Review & Editing, J.O.O., M.M.D., E.L.L., P.M.K, S.A.M., N.G., J.F., G.A.E.M., R.N., L.R.N., Y.B., and M.C.A.O.; Visualization, E.L.L., and N.G.; Supervision, J.O.O., R.G.E., L.E., M.M.D., E.M., M.H.M., S.E., D.N., P.M.K, S.K., V.J.B., Y.E., G.A.E.M., R.N., L.R.N., Y.B., and M.C.A.O.; Project Administration, J.O.O., R.G.E., M.M.D., S.E., G.A.E.M., R.N., L.R.N., Y.B., and M.C.A.O.; Funding Acquisition, J.O.O. All authors read and approved the final manuscript.
ACKNOWLEDGMENTS:
We gratefully acknowledge Dr. Manaouda Malachie, Minister of Health of Cameroon; Dr. Phanuel Habimana, WHO Cameroon Country Representative; Dr. Thierno Balde, WHO AFRO COVID-19 Regional Incident Manager; and Dr. Abdou Salam Gueye, WHO AFRO Regional Emergency Director, for their invaluable support. We extend our appreciation to Professor Bright Adu, Coordinator of the Noguchi NGS and WHO-Africa CDC SARS-CoV-2 Regional Sequencing Laboratory, and Professor Edyth Parker, Department of Immunology and Microbiology, The Scripps Research Institute, for their expertise in phylogenetic analysis and bioinformatics. We thank Dr. Safa Boujemaa from Biologica Training and Consulting Group for her thorough manuscript review. Our gratitude extends to the laboratory technicians at the National Public Health Laboratory and the Centre Pasteur du Cameroun, as well as to Dr. Amadou Diallo, Mr. Moise Christian Meka, Mr. Stéphane Tewo, Mr. Christian Mouangue, Dr. Jayne Byakika Tusiime, and Dr. Humphrey Cyprian Karamagi for their significant contributions.
References
- Esso L, Epée E, Bilounga C, Abah A, Hamadou A, Dibongue E, et al. Cameroon’s bold response to the COVID-19 pandemic during the first and second waves. Lancet Infect Dis. 2021; 21(11): 1484-1485.
- Njouom R, Sadeuh-Mba SA, Tchatchueng J, Diagne MM, Dia N, Tagnouokam PAN, et al. Coding-complete genome sequence and phylogenetic relatedness of a SARS-CoV-2 strain detected in March 2020 in Cameroon. Microbiol Resour Announc. 2021;10(13):e00093-21.
- Oude Munnink BB, Worp N, Nieuwenhuijse DF, Sikkema RS, Haagmans B, Fouchier RAM, et al. The next phase of SARS-CoV-2 surveillance: real-time molecular epidemiology. Nat Med. 2021;27(9):1518-1524.
- Tegally H, Moir M, Everatt J, Giovanetti M, Scheepers C, Wilkinson E, et al. Continued emergence and evolution of Omicron in South Africa: new BA.4 and BA.5 lineages. Nat Med. 2022;28(9):1785-1790.
- Wolter N, Jassat W, Walaza S, Welch R, Moultrie H, Groome MJ, et al. Clinical severity of SARS-CoV-2 Omicron BA.4 and BA.5 lineages compared to BA.1 and Delta in South Africa. Nat Commun. 2022; 13(1): 5860.
- Islam MR, Shahriar M, Bhuiyan MA. The latest Omicron BA.4 and BA.5 lineages are frowning toward COVID‐19 preventive measures: a threat to global public health. Health Sci Rep. 2022;5(6):e884.
- Lo CC, Shakya M, Connor R, Davenport K, Flynn M, Gutiérrez AM, et al. EDGE COVID-19: a web platform to generate submission-ready genomes from SARS-CoV-2 sequencing efforts. Bioinformatics. 2022; 38(12): 3254-3256.
- Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, et al. BA.2.12.1, BA.4, and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022; 608(7923): 593-602.
- Fonager J, Bennedbak M, Bager P, Wohlfahrt J, Ellegaard KM, Ingham AC, et al. Molecular epidemiology of the SARS-CoV-2 variant Omicron BA.2 sub-lineage in Denmark, 29 November 2021 to 2 January 2022. Euro Surveill. 2022; 27(10): 2200181.
- World Health Organization. COVID-19 Surveillance Guidance: Global Surveillance for COVID-19 Caused by Human Infection with COVID-19 Virus. WHO; 2022.
- Fokam J, Nka AD, Teto G, Beloumou G, Dambaya B, Kamgaing N, et al. XBB.1, BQ1.1, and atypical BA.4.6/XBB.1 recombinants predominate current SARS-CoV-2 wavelets with flu-like symptoms in Cameroon: a snapshot from genomic surveillance. Int J Infect Dis. 2024; 139: 154-157.
- World Health Organization. Statement on the Fifteenth Meeting of the International Health Regulations (2005) Emergency Committee Regarding the Coronavirus Disease (COVID-19) Pandemic. WHO; 2023.
- Khoury DS, Docken SS, Subbarao K, Kent SJ, Davenport MP. The shifting sands of COVID-19 immunity: endemic cross-reactive immunity against new SARS-CoV-2 variants. Nat Rev Immunol. 2023; 23(5): 279-290.
- Morens DM, Taubenberger JK, Fauci AS. Universal coronavirus vaccines — an urgent need. N Engl J Med. 2022; 386(4): 297-299.
- Bhoyar RC, Jain A, Sehgal P, Divakar MK, Sharma D, Imran M, et al. High-throughput detection and genetic epidemiology of SARS-CoV-2 using COVIDSeq next-generation sequencing. PLoS One. 2021; 16(2): e0247115.
- Illumina Inc. COVIDSeq Test Reference Guide. Document #1000000126053 v06. Illumina; 2021.
- Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021; 38(7): 3022-3027.
- Thompson RN, Hill EM, Gog JR. SARS-CoV-2 epidemiology: a review of major drivers and analytical approaches. Nat Rev Microbiol. 2023; 21(1): 46-60.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25(16): 2078-2079.
- Lo CC, Chain PSG. Rapid evaluation and quality control of next-generation sequencing data with FaQCs. BMC Bioinformatics. 2014; 15(1): 366.
- O’Toole Á, Scher E, Underwood A, Jackson B, Hill V, McCrone JT, et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021; 7(2): veab064.
- Rambaut A, Holmes EC, O’Toole Á, Hill V, McCrone JT, Ruis C, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020; 5(11): 1403-1407.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020; 37(5): 1530-1534.
- Yu G, Smith DK, Zhu H, Guan Y, Lam TT. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017; 8(1): 28-36.
- Jackson B, Boni MF, Bull MJ, Colleran A, Colquhoun RM, Darby AC, et al. Generation and transmission of interlineage recombinants in the SARS-CoV-2 pandemic. Cell. 2021; 184(20): 5179-5188.
- Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018; 4(1): vey016.
- Oude Munnink BB, Koopmans MPG. Advanced molecular surveillance strategies for respiratory viruses: lessons from SARS-CoV-2. Nat Rev Microbiol. 2024; 22(1): 15-25.
- Mlcochova P, Kemp SA, Gupta RK. SARS-CoV-2 variant evolution in 2023: impact on diagnostics and therapeutics. Nat Rev Genet. 2024; 25(1): 114-119.
- Ntoumi F, Vouvoungui JC, Ibara BR, Akiana J, Moukassa D. Strengthening COVID-19 surveillance in Central Africa: a 2023-2024 analysis. Int J Infect Dis. 2024; 128: 215-221.
- Inzaule SC, Tessema SK, Kebede Y, Ogwell Ouma AE, Nkengasong JN. Next-generation sequencing in African SARS-CoV-2 surveillance: 2024 update. Lancet Microbe. 2024; 5(2): e70-e78.
- Abdool Karim SS, de Oliveira T. SARS-CoV-2 surveillance strategies for 2024: African perspective. N Engl J Med. 2024; 385(2): 166-168.
- Happi AN, Nkengasong JN. Three years of COVID-19 in Africa: 2024 update. Nature. 2024; 615(7891): 22-25.
- Bedford T, Hodcroft EB, Neher RA, Rambaut A, Pybus OG, Kellam P. Viral genome surveillance: new horizons in understanding SARS-CoV-2 evolution. Nat Rev Genet. 2024; 25(2): 223-235.

